
A retrovirus is a type of virus that inserts a copy of its RNA genome into the DNA of a host cell that it invades, thus changing the genome of that cell. Once inside the host cell's cytoplasm, the virus uses its own reverse transcriptase enzyme to produce DNA from its RNA genome, the reverse of the usual pattern, thus retro (backwards). The new DNA is then incorporated into the host cell genome by an integrase enzyme, at which point the retroviral DNA is referred to as a provirus. The host cell then treats the viral DNA as part of its own genome, transcribing and translating the viral genes along with the cell's own genes, producing the proteins required to assemble new copies of the virus.

Poliovirus, the causative agent of polio, is a serotype of the species Enterovirus C, in the family of Picornaviridae.

Picornaviruses are a group of related nonenveloped RNA viruses which infect vertebrates including mammals and birds. They are viruses that represent a large family of small, positive-sense, single-stranded RNA viruses with a 30-nm icosahedral capsid. The viruses in this family can cause a range of diseases including the common cold, poliomyelitis, meningitis, hepatitis, and paralysis.

Gammaretrovirus is a genus in the Retroviridae family. Example species are the murine leukemia virus and the feline leukemia virus. They cause various sarcomas, leukemias and immune deficiencies in mammals, reptiles and birds.
The genome and proteins of HIV have been the subject of extensive research since the discovery of the virus in 1983. "In the search for the causative agent, it was initially believed that the virus was a form of the Human T-cell leukemia virus (HTLV), which was known at the time to affect the human immune system and cause certain leukemias. However, researchers at the Pasteur Institute in Paris isolated a previously unknown and genetically distinct retrovirus in patients with AIDS which was later named HIV." Each virion comprises a viral envelope and associated matrix enclosing a capsid, which itself encloses two copies of the single-stranded RNA genome and several enzymes. The discovery of the virus itself occurred two years following the report of the first major cases of AIDS-associated illnesses.
The murine leukemia viruses are retroviruses named for their ability to cause cancer in murine (mouse) hosts. Some MLVs may infect other vertebrates. MLVs include both exogenous and endogenous viruses. Replicating MLVs have a positive sense, single-stranded RNA (ssRNA) genome that replicates through a DNA intermediate via the process of reverse transcription.
The Simian foamy virus (SFV) is a species of the genus Spumavirus, which belongs to the family of Retroviridae. It has been identified in a wide variety of primates, including pro-simians, New World and Old World monkeys as well as apes, and each species has been shown to harbor a unique (species-specific) strain of SFV, including African green monkeys, baboons, macaques and chimpanzees. As it is related to the more well-known retrovirus human immunodeficiency virus (HIV), its discovery in primates has led to some speculation that HIV may have been spread to the human species in Africa through contact with blood from apes, monkeys, and other primates, most likely through bushmeat hunting practices.

A long terminal repeat (LTR) is a pair of identical sequences of DNA, several hundred base pairs long, which occur in eukaryotic genomes on either end of a series of genes or pseudogenes that form a retrotransposon or an endogenous retrovirus or a retroviral provirus. All retroviral genomes are flanked by LTRs, while there are some retrotransposons without LTRs. Typically, an element flanked by a pair of LTRs will encode a reverse transcriptase and an integrase, allowing the element to be copied and inserted at a different location of the genome. Copies of such an LTR-flanked element can often be found hundreds or thousands of times in a genome. LTR retrotransposons comprise about 8% of the human genome.
Crinivirus, formerly the lettuce infectious yellows virus group, is a genus of viruses, in the family Closteroviridae. They are linear, single-stranded positive sense RNA viruses. There are 14 species in this genus. Diseases associated with this genus include: yellowing and necrosis, particularly affecting the phloem.

The Citrus tristeza virus replication signal is a regulatory element involved in a viral replication signal which is highly conserved in citrus tristeza viruses. Replication signals are required for viral replication and are usually found near the 5' and 3' termini of protein coding genes. This element is predicted to form ten stem loop structures some of which are essential for functions that provide for efficient viral replication.
The Coronavirus 3′ stem-loop II-like motif is a secondary structure motif identified in the 3′ untranslated region (3′UTR) of astrovirus, coronavirus and equine rhinovirus genomes. Its function is unknown, but various viral 3′ UTR regions have been found to play roles in viral replication and packaging.

The Coronavirus packaging signal is a conserved cis-regulatory element found in Betacoronavirus. It has an important role in regulating the packaging of the viral genome into the capsid. As part of the viral life cycle, within the infected cell, the viral genome becomes associated with viral proteins and assembles into new infective progeny viruses. This process is called packaging and is vital for viral replication.

The Gammaretrovirus core encapsidation signal is an RNA element known to be essential for stable dimerisation and efficient genome packaging during virus assembly. Dimerisation of the viral RNA genomes is proposed to act as an RNA conformational switch which exposes conserved UCUG elements and enables efficient genome encapsidation. The structure of this element is composed of three stem-loops. Two of the stem-loops called SL-C and SL-D form a single co-axial extend helix.

The retroviral psi packaging element, also known as the Ψ RNA packaging signal, is a cis-acting RNA element identified in the genomes of the retroviruses Human immunodeficiency virus (HIV) and Simian immunodeficiency virus (SIV). It is involved in regulating the essential process of packaging the retroviral RNA genome into the viral capsid during replication. The final virion contains a dimer of two identical unspliced copies of the viral genome.

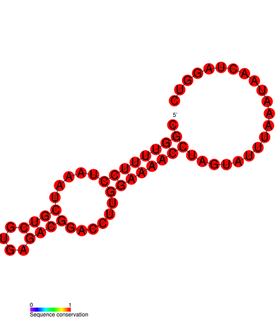
The HBV RNA encapsidation signal epsilon is an element essential for HBV virus replication.
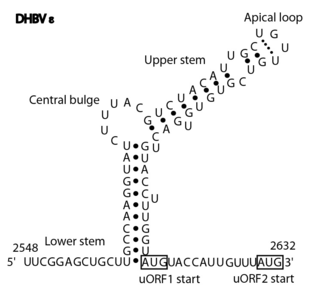
The Avian HBV RNA encapsidation signal epsilon is an RNA structure that is shown to facilitate encapsidation of the pregenomic RNA required for viral replication. There are two main classes of encapsidation signals in avian hepatitis B viruses - Duck hepatitis B virus (DHBV) and Heron hepatitis B virus (HHBV) like. DHBV is used as a model to understand human Hepatitis B virus. Although studies have shown that the HHBV epsilon has less pairing in the upper stem than DHBV, this pairing is not absolutely required for DHBV infection in ducks.
Mason-Pfizer monkey virus (M-PMV), formerly Simian retrovirus (SRV), is a species of retroviruses that usually infect and cause a fatal immune deficiency in Asian macaques. The ssRNA virus appears sporadically in mammary carcinoma of captive macaques at breeding facilities which expected as the natural host, but the prevalence of this virus in feral macaques remains unknown. M-PMV was transmitted naturally by virus-containing body fluids, via biting, scratching, grooming, and fighting. Cross contaminated instruments or equipment (fomite) can also spread this virus among animals.
Bovine foamy virus (BFV) is a ss(+)RNA retrovirus that belongs to the genus spumaviridae. Spumaviruses differ from the other six members of family retroviridae, both structurally and in pathogenic nature. Spumaviruses derive their name from spuma the latin for "foam". The 'foam' aspect of 'foamy virus' comes from syncytium formation and the rapid vacuolization of infected cells, creating a 'foamy' appearance.
Coronavirus genomes are positive-sense single-stranded RNA molecules with an untranslated region (UTR) at the 5′ end which is called the 5′ UTR. The 5′ UTR is responsible for important biological functions, such as viral replication, transcription and packaging. The 5′ UTR has a conserved RNA secondary structure but different Coronavirus genera have different structural features described below.

Flavivirus 5' UTR are untranslated regions in the genome of viruses in the genus Flavivirus.














