An intron is any nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word intron is derived from the term intragenic region, i.e., a region inside a gene. The term intron refers to both the DNA sequence within a gene and the corresponding RNA sequence in RNA transcripts. The non-intron sequences that become joined by this RNA processing to form the mature RNA are called exons.

RNA splicing is a process in molecular biology where a newly-made precursor messenger RNA (pre-mRNA) transcript is transformed into a mature messenger RNA (mRNA). It works by removing all the introns and splicing back together exons. For nuclear-encoded genes, splicing occurs in the nucleus either during or immediately after transcription. For those eukaryotic genes that contain introns, splicing is usually needed to create an mRNA molecule that can be translated into protein. For many eukaryotic introns, splicing occurs in a series of reactions which are catalyzed by the spliceosome, a complex of small nuclear ribonucleoproteins (snRNPs). There exist self-splicing introns, that is, ribozymes that can catalyze their own excision from their parent RNA molecule. The process of transcription, splicing and translation is called gene expression, the central dogma of molecular biology.

A spliceosome is a large ribonucleoprotein (RNP) complex found primarily within the nucleus of eukaryotic cells. The spliceosome is assembled from small nuclear RNAs (snRNA) and numerous proteins. Small nuclear RNA (snRNA) molecules bind to specific proteins to form a small nuclear ribonucleoprotein complex, which in turn combines with other snRNPs to form a large ribonucleoprotein complex called a spliceosome. The spliceosome removes introns from a transcribed pre-mRNA, a type of primary transcript. This process is generally referred to as splicing. An analogy is a film editor, who selectively cuts out irrelevant or incorrect material from the initial film and sends the cleaned-up version to the director for the final cut.
snRNPs, or small nuclear ribonucleoproteins, are RNA-protein complexes that combine with unmodified pre-mRNA and various other proteins to form a spliceosome, a large RNA-protein molecular complex upon which splicing of pre-mRNA occurs. The action of snRNPs is essential to the removal of introns from pre-mRNA, a critical aspect of post-transcriptional modification of RNA, occurring only in the nucleus of eukaryotic cells. Additionally, U7 snRNP is not involved in splicing at all, as U7 snRNP is responsible for processing the 3′ stem-loop of histone pre-mRNA.
Small nuclear RNA (snRNA) is a class of small RNA molecules that are found within the splicing speckles and Cajal bodies of the cell nucleus in eukaryotic cells. The length of an average snRNA is approximately 150 nucleotides. They are transcribed by either RNA polymerase II or RNA polymerase III. Their primary function is in the processing of pre-messenger RNA (hnRNA) in the nucleus. They have also been shown to aid in the regulation of transcription factors or RNA polymerase II, and maintaining the telomeres.

The minor spliceosome is a ribonucleoprotein complex that catalyses the removal (splicing) of an atypical class of spliceosomal introns (U12-type) from messenger RNAs in some clades of eukaryotes. This process is called noncanonical splicing, as opposed to U2-dependent canonical splicing. U12-type introns represent less than 1% of all introns in human cells. However they are found in genes performing essential cellular functions.
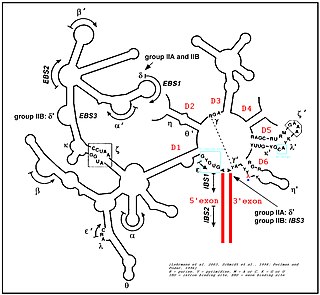
Group II introns are a large class of self-catalytic ribozymes and mobile genetic elements found within the genes of all three domains of life. Ribozyme activity can occur under high-salt conditions in vitro. However, assistance from proteins is required for in vivo splicing. In contrast to group I introns, intron excision occurs in the absence of GTP and involves the formation of a lariat, with an A-residue branchpoint strongly resembling that found in lariats formed during splicing of nuclear pre-mRNA. It is hypothesized that pre-mRNA splicing may have evolved from group II introns, due to the similar catalytic mechanism as well as the structural similarity of the Group II Domain V substructure to the U6/U2 extended snRNA. Finally, their ability to site-specifically insert into DNA sites has been exploited as a tool for biotechnology. For example, group II introns can be modified to make site-specific genome insertions and deliver cargo DNA such as reporter genes or lox sites
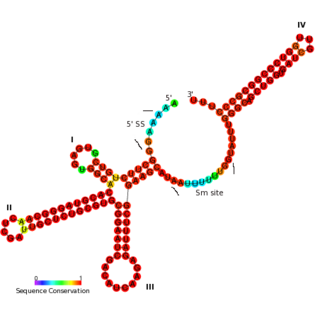
The U11 snRNA is an important non-coding RNA in the minor spliceosome protein complex, which activates the alternative splicing mechanism. The minor spliceosome is associated with similar protein components as the major spliceosome. It uses U11 snRNA to recognize the 5' splice site while U12 snRNA binds to the branchpoint to recognize the 3' splice site.

U1 spliceosomal RNA is the small nuclear RNA (snRNA) component of U1 snRNP, an RNA-protein complex that combines with other snRNPs, unmodified pre-mRNA, and various other proteins to assemble a spliceosome, a large RNA-protein molecular complex upon which splicing of pre-mRNA occurs. Splicing, or the removal of introns, is a major aspect of post-transcriptional modification, and takes place only in the nucleus of eukaryotes.

U2 spliceosomal snRNAs are a species of small nuclear RNA (snRNA) molecules found in the major spliceosomal (Sm) machinery of virtually all eukaryotic organisms. In vivo, U2 snRNA along with its associated polypeptides assemble to produce the U2 small nuclear ribonucleoprotein (snRNP), an essential component of the major spliceosomal complex. The major spliceosomal-splicing pathway is occasionally referred to as U2 dependent, based on a class of Sm intron—found in mRNA primary transcripts—that are recognized exclusively by the U2 snRNP during early stages of spliceosomal assembly. In addition to U2 dependent intron recognition, U2 snRNA has been theorized to serve a catalytic role in the chemistry of pre-RNA splicing as well. Similar to ribosomal RNAs (rRNAs), Sm snRNAs must mediate both RNA:RNA and RNA:protein contacts and hence have evolved specialized, highly conserved, primary and secondary structural elements to facilitate these types of interactions.

The U4 small nuclear Ribo-Nucleic Acid is a non-coding RNA component of the major or U2-dependent spliceosome – a eukaryotic molecular machine involved in the splicing of pre-messenger RNA (pre-mRNA). It forms a duplex with U6, and with each splicing round, it is displaced from the U6 snRNA in an ATP-dependent manner, allowing U6 to re-fold and create the active site for splicing catalysis. A recycling process involving protein Brr2 releases U4 from U6, while protein Prp24 re-anneals U4 and U6. The crystal structure of a 5′ stem-loop of U4 in complex with a binding protein has been solved.

U6 snRNA is the non-coding small nuclear RNA (snRNA) component of U6 snRNP, an RNA-protein complex that combines with other snRNPs, unmodified pre-mRNA, and various other proteins to assemble a spliceosome, a large RNA-protein molecular complex that catalyzes the excision of introns from pre-mRNA. Splicing, or the removal of introns, is a major aspect of post-transcriptional modification and takes place only in the nucleus of eukaryotes.

Pre-mRNA-processing-splicing factor 8 is a protein that in humans is encoded by the PRPF8 gene.

PRP31 pre-mRNA processing factor 31 homolog , also known as PRPF31, is a protein which in humans is encoded by the PRPF31 gene.
U4/U6 small nuclear ribonucleoprotein Prp3 is a protein that in humans is encoded by the PRPF3 gene.

Prp24 is a protein part of the pre-messenger RNA splicing process and aids the binding of U6 snRNA to U4 snRNA during the formation of spliceosomes. Found in eukaryotes from yeast to E. coli, fungi, and humans, Prp24 was initially discovered to be an important element of RNA splicing in 1989. Mutations in Prp24 were later discovered in 1991 to suppress mutations in U4 that resulted in cold-sensitive strains of yeast, indicating its involvement in the reformation of the U4/U6 duplex after the catalytic steps of splicing.
Numerous key discoveries in biology have emerged from studies of RNA, including seminal work in the fields of biochemistry, genetics, microbiology, molecular biology, molecular evolution, and structural biology. As of 2010, 30 scientists have been awarded Nobel Prizes for experimental work that includes studies of RNA. Specific discoveries of high biological significance are discussed in this article.
Messenger RNP is mRNA with bound proteins. mRNA does not exist "naked" in vivo but is always bound by various proteins while being synthesized, spliced, exported, and translated in the cytoplasm.
Ring Finger Protein 113A is a protein that in humans is encoded by the RNF113A gene. It is found in humans on the X Chromosome. RNF113A contains two highly conserved domains, the RING finger domain and Zinc finger domain. RING finger domains have been associated with some tumor suppressors and cytokine receptor-associated molecules. These domains also act in DNA repair and mediating protein-protein interactions. Aliases of RNF113A across taxa include RNF113, CWC24, and ZNF183.

Prp8 refers to both the Prp8 protein and Prp8 gene. Prp8's name originates from its involvement in pre-mRNA processing. The Prp8 protein is a large, highly conserved, and unique protein that resides in the catalytic core of the spliceosome and has been found to have a central role in molecular rearrangements that occur there. Prp8 protein is a major central component of the catalytic core in the spliceosome, and the spliceosome is responsible for splicing of precursor mRNA that contains introns and exons. Unexpressed introns are removed by the spliceosome complex in order to create a more concise mRNA transcript. Splicing is just one of many different post-transcriptional modifications that mRNA must undergo before translation. Prp8 has also been hypothesized to be a cofactor in RNA catalysis.














