A bacterial artificial chromosome (BAC) is a DNA construct, based on a functional fertility plasmid, used for transforming and cloning in bacteria, usually E. coli. F-plasmids play a crucial role because they contain partition genes that promote the even distribution of plasmids after bacterial cell division. The bacterial artificial chromosome's usual insert size is 150–350 kbp. A similar cloning vector called a PAC has also been produced from the DNA of P1 bacteriophage.

A cloning vector is a small piece of DNA that can be stably maintained in an organism, and into which a foreign DNA fragment can be inserted for cloning purposes. The cloning vector may be DNA taken from a virus, the cell of a higher organism, or it may be the plasmid of a bacterium. The vector contains features that allow for the convenient insertion of a DNA fragment into the vector or its removal from the vector, for example through the presence of restriction sites. The vector and the foreign DNA may be treated with a restriction enzyme that cuts the DNA, and DNA fragments thus generated contain either blunt ends or overhangs known as sticky ends, and vector DNA and foreign DNA with compatible ends can then be joined by molecular ligation. After a DNA fragment has been cloned into a cloning vector, it may be further subcloned into another vector designed for more specific use.

Yeast artificial chromosomes (YACs) are genetically engineered chromosomes derived from the DNA of the yeast, Saccharomyces cerevisiae, which is then ligated into a bacterial plasmid. By inserting large fragments of DNA, from 100–1000 kb, the inserted sequences can be cloned and physically mapped using a process called chromosome walking. This is the process that was initially used for the Human Genome Project, however due to stability issues, YACs were abandoned for the use of bacterial artificial chromosome
Repeated sequences are short or long patterns of nucleic acids that occur in multiple copies throughout the genome. In many organisms, a significant fraction of the genomic DNA is repetitive, with over two-thirds of the sequence consisting of repetitive elements in humans. Some of these repeated sequences are necessary for maintaining important genome structures such as telomeres or centromeres.
Forward genetics is a molecular genetics approach of determining the genetic basis responsible for a phenotype. Forward genetics provides an unbiased approach because it relies heavily on identifying the genes or genetic factors that cause a particular phenotype or trait of interest.
Primer walking is a technique used to clone a gene from its known closest markers. As a result, it is employed in cloning and sequencing efforts in plants, fungi, and mammals with minor alterations. This technique, also known as "directed sequencing," employs a series of Sanger sequencing reactions to either confirm the reference sequence of a known plasmid or PCR product based on the reference sequence or to discover the unknown sequence of a full plasmid or PCR product by designing primers to sequence overlapping sections.
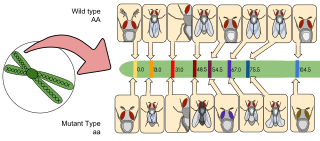
Gene mapping or genome mapping describes the methods used to identify the location of a gene on a chromosome and the distances between genes. Gene mapping can also describe the distances between different sites within a gene.
A genomic library is a collection of overlapping DNA fragments that together make up the total genomic DNA of a single organism. The DNA is stored in a population of identical vectors, each containing a different insert of DNA. In order to construct a genomic library, the organism's DNA is extracted from cells and then digested with a restriction enzyme to cut the DNA into fragments of a specific size. The fragments are then inserted into the vector using DNA ligase. Next, the vector DNA can be taken up by a host organism - commonly a population of Escherichia coli or yeast - with each cell containing only one vector molecule. Using a host cell to carry the vector allows for easy amplification and retrieval of specific clones from the library for analysis.
Fosmids are similar to cosmids but are based on the bacterial F-plasmid. The cloning vector is limited, as a host can only contain one fosmid molecule. Fosmids can hold DNA inserts of up to 40 kb in size; often the source of the insert is random genomic DNA. A fosmid library is prepared by extracting the genomic DNA from the target organism and cloning it into the fosmid vector. The ligation mix is then packaged into phage particles and the DNA is transfected into the bacterial host. Bacterial clones propagate the fosmid library. The low copy number offers higher stability than vectors with relatively higher copy numbers, including cosmids. Fosmids may be useful for constructing stable libraries from complex genomes. Fosmids have high structural stability and have been found to maintain human DNA effectively even after 100 generations of bacterial growth. Fosmid clones were used to help assess the accuracy of the Public Human Genome Sequence.
In the fields of bioinformatics and computational biology, Genome survey sequences (GSS) are nucleotide sequences similar to expressed sequence tags (ESTs) that the only difference is that most of them are genomic in origin, rather than mRNA.
Artificial gene synthesis, or simply gene synthesis, refers to a group of methods that are used in synthetic biology to construct and assemble genes from nucleotides de novo. Unlike DNA synthesis in living cells, artificial gene synthesis does not require template DNA, allowing virtually any DNA sequence to be synthesized in the laboratory. It comprises two main steps, the first of which is solid-phase DNA synthesis, sometimes known as DNA printing. This produces oligonucleotide fragments that are generally under 200 base pairs. The second step then involves connecting these oligonucleotide fragments using various DNA assembly methods. Because artificial gene synthesis does not require template DNA, it is theoretically possible to make a completely synthetic DNA molecule with no limits on the nucleotide sequence or size.
A P1-derived artificial chromosome, or PAC, is a DNA construct derived from the DNA of P1 bacteriophages and Bacterial artificial chromosome. It can carry large amounts of other sequences for a variety of bioengineering purposes in bacteria. It is one type of the efficient cloning vector used to clone DNA fragments in Escherichia coli cells.
Paired-end tags (PET) are the short sequences at the 5’ and 3' ends of a DNA fragment which are unique enough that they (theoretically) exist together only once in a genome, therefore making the sequence of the DNA in between them available upon search or upon further sequencing. Paired-end tags (PET) exist in PET libraries with the intervening DNA absent, that is, a PET "represents" a larger fragment of genomic or cDNA by consisting of a short 5' linker sequence, a short 5' sequence tag, a short 3' sequence tag, and a short 3' linker sequence. It was shown conceptually that 13 base pairs are sufficient to map tags uniquely. However, longer sequences are more practical for mapping reads uniquely. The endonucleases used to produce PETs give longer tags but sequences of 50–100 base pairs would be optimal for both mapping and cost efficiency. After extracting the PETs from many DNA fragments, they are linked (concatenated) together for efficient sequencing. On average, 20–30 tags could be sequenced with the Sanger method, which has a longer read length. Since the tag sequences are short, individual PETs are well suited for next-generation sequencing that has short read lengths and higher throughput. The main advantages of PET sequencing are its reduced cost by sequencing only short fragments, detection of structural variants in the genome, and increased specificity when aligning back to the genome compared to single tags, which involves only one end of the DNA fragment.

Molecular cloning is a set of experimental methods in molecular biology that are used to assemble recombinant DNA molecules and to direct their replication within host organisms. The use of the word cloning refers to the fact that the method involves the replication of one molecule to produce a population of cells with identical DNA molecules. Molecular cloning generally uses DNA sequences from two different organisms: the species that is the source of the DNA to be cloned, and the species that will serve as the living host for replication of the recombinant DNA. Molecular cloning methods are central to many contemporary areas of modern biology and medicine.
A mega-telomere, is an extremely long telomere sequence that sits on the end of chromosomes and prevents the loss of genetic information during cell replication. Like regular telomeres, mega-telomeres are made of a repetitive sequence of DNA and associated proteins, and are located on the ends of chromosomes. However, mega-telomeres are substantially longer than regular telomeres, ranging in size from 50 kilobases to several megabases.

Jumping libraries or junction-fragment libraries are collections of genomic DNA fragments generated by chromosome jumping. These libraries allow the analysis of large areas of the genome and overcome distance limitations in common cloning techniques. A jumping library clone is composed of two stretches of DNA that are usually located many kilobases away from each other. The stretch of DNA located between these two "ends" is deleted by a series of biochemical manipulations carried out at the start of this cloning technique.
Synthetic genome is a synthetically built genome whose formation involves either genetic modification on pre-existing life forms or artificial gene synthesis to create new DNA or entire lifeforms. The field that studies synthetic genomes is called synthetic genomics.
Physical map is a technique used in molecular biology to find the order and physical distance between DNA base pairs by DNA markers. It is one of the gene mapping techniques which can determine the sequence of DNA base pairs with high accuracy. Genetic mapping, another approach of gene mapping, can provide markers needed for the physical mapping. However, as the former deduces the relative gene position by recombination frequencies, it is less accurate than the latter.

Vectorette PCR is a variation of polymerase chain reaction (PCR) designed in 1988. The original PCR was created and also patented during the 1980s. Vectorette PCR was first noted and described in an article in 1990 by John H. Riley and his team. Since then, multiple variants of PCR have been created. Vectorette PCR focuses on amplifying a specific sequence obtained from an internal sequence that is originally known until the fragment end. Multiple researches have taken this method as an opportunity to conduct experiments in order to uncover the potential uses that can be derived from Vectorette PCR.
Long-range restriction mapping is an alternative genomic mapping technique to short-range, also called fine-scale mapping. Both forms utilize restriction enzymes in order to decipher the previously unknown order of DNA segments; the main difference between the two being the amount of DNA that comprises the final map. The unknown DNA is broken into many smaller fragments by these restriction enzymes at specific sites on the molecule, and then the fragments can later be analyzed by their individual sizes. A final long-range map can span hundreds to thousands of kilobytes of genetic data at many different loci.







