 | |
| Names | |
|---|---|
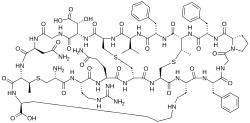
| IUPAC name (1S,4S,13S,16S,19R,22S,25S,28R,31S,37S,41R,44R,47S,50S,53R,56R,65S)-44-amino-37-(2-amino-2-oxoethyl)-50-(3-amino-3-oxopropyl)-4,16,22-tribenzyl-47-(3-carbamimidamidopropyl)-31-[(R)-carboxy(hydroxy)methyl]-41,70-dimethyl-2,5,8,14,17,20,23,26,29,32,35,38,45,48,51,54,57,67-octadecaoxo-25-propan-2-yl-42,69,72-trithia-3,6,9,15,18,21,24,27,30,33,36,39,46,49,52,55,58,60,66-nonadecazapentacyclo[38.18.9.319,56.328,53.09,13]triheptacontane-65-carboxylic acid | |
| Other names Lanthiopeptin; NSC-71936; Ro09-198 | |
| Identifiers | |
3D model (JSmol) | |
| ChEBI | |
| ChemSpider | |
PubChem CID | |
| UNII | |
| |
| |
| Properties | |
| C89H125N25O25S3 | |
| Molar mass | 2041.31 g·mol−1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
Cinnamycin is a tetracyclic antibacterial peptide produced by Streptomyces cinnamoneus containing 19 amino acid residues including the unusual amino acids threo-3-methyl-lanthionine, meso-lanthionine, lysinoalanine, and 3-hydroxyaspartic acid.
Contents
- Structure
- Receptor–molecule interactions
- Cinnamycin gene cluster – Cin
- Biosynthesis
- Immunity mechanism
- Cinnamycin-like compounds
- Biological activities
- References
Cinnamycin belongs to the class of molecules known as lantibiotics which belongs to ribosomally synthesized post-translationally modified peptides. The unique receptor for cinnamycin is phosphatidylethanolamine (PE) lipids which is a major compound present in many bacterial cell membranes.
Cinnamycin was first isolated in 1952 and some other compounds with similar sequence and structure were found later. [1]