
Protein biosynthesis is a core biological process, occurring inside cells, balancing the loss of cellular proteins through the production of new proteins. Proteins perform a number of critical functions as enzymes, structural proteins or hormones. Protein synthesis is a very similar process for both prokaryotes and eukaryotes but there are some distinct differences.
Peptide nucleic acid (PNA) is an artificially synthesized polymer similar to DNA or RNA.
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, research, and forensics. Commonly made in the laboratory by solid-phase chemical synthesis, these small bits of nucleic acids can be manufactured as single-stranded molecules with any user-specified sequence, and so are vital for artificial gene synthesis, polymerase chain reaction (PCR), DNA sequencing, molecular cloning and as molecular probes. In nature, oligonucleotides are usually found as small RNA molecules that function in the regulation of gene expression, or are degradation intermediates derived from the breakdown of larger nucleic acid molecules.
Gene silencing is the regulation of gene expression in a cell to prevent the expression of a certain gene. Gene silencing can occur during either transcription or translation and is often used in research. In particular, methods used to silence genes are being increasingly used to produce therapeutics to combat cancer and other diseases, such as infectious diseases and neurodegenerative disorders.
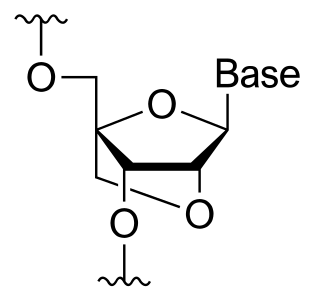
A locked nucleic acid (LNA), also known as bridged nucleic acid (BNA), and often referred to as inaccessible RNA, is a modified RNA nucleotide in which the ribose moiety is modified with an extra bridge connecting the 2' oxygen and 4' carbon. The bridge "locks" the ribose in the 3'-endo (North) conformation, which is often found in the A-form duplexes. This structure can be attributed to the increased stability against enzymatic degradation; moreover the structure of LNA has improved specificity and affinity as a monomer or a constituent of an oligonucleotide. LNA nucleotides can be mixed with DNA or RNA residues in the oligonucleotide, in effect hybridizing with DNA or RNA according to Watson-Crick base-pairing rules.
Site-directed mutagenesis is a molecular biology method that is used to make specific and intentional changes to the DNA sequence of a gene and any gene products. Also called site-specific mutagenesis or oligonucleotide-directed mutagenesis, it is used for investigating the structure and biological activity of DNA, RNA, and protein molecules, and for protein engineering.

Small interfering RNA (siRNA), sometimes known as short interfering RNA or silencing RNA, is a class of double-stranded RNA non-coding RNA molecules, typically 20-24 base pairs in length, similar to miRNA, and operating within the RNA interference (RNAi) pathway. It interferes with the expression of specific genes with complementary nucleotide sequences by degrading mRNA after transcription, preventing translation.

A frameshift mutation is a genetic mutation caused by indels of a number of nucleotides in a DNA sequence that is not divisible by three. Due to the triplet nature of gene expression by codons, the insertion or deletion can change the reading frame, resulting in a completely different translation from the original. The earlier in the sequence the deletion or insertion occurs, the more altered the protein. A frameshift mutation is not the same as a single-nucleotide polymorphism in which a nucleotide is replaced, rather than inserted or deleted. A frameshift mutation will in general cause the reading of the codons after the mutation to code for different amino acids. The frameshift mutation will also alter the first stop codon encountered in the sequence. The polypeptide being created could be abnormally short or abnormally long, and will most likely not be functional.

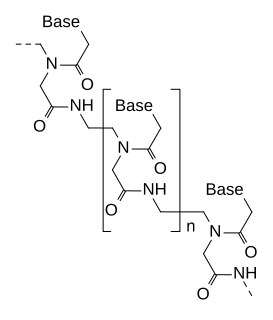
A Morpholino, also known as a Morpholino oligomer and as a phosphorodiamidate Morpholino oligomer (PMO), is a type of oligomer molecule used in molecular biology to modify gene expression. Its molecular structure contains DNA bases attached to a backbone of methylenemorpholine rings linked through phosphorodiamidate groups. Morpholinos block access of other molecules to small specific sequences of the base-pairing surfaces of ribonucleic acid (RNA). Morpholinos are used as research tools for reverse genetics by knocking down gene function.

Antisense RNA (asRNA), also referred to as antisense transcript, natural antisense transcript (NAT) or antisense oligonucleotide, is a single stranded RNA that is complementary to a protein coding messenger RNA (mRNA) with which it hybridizes, and thereby blocks its translation into protein. asRNAs have been found in both prokaryotes and eukaryotes, and can be classified into short and long non-coding RNAs (ncRNAs). The primary function of asRNA is regulating gene expression. asRNAs may also be produced synthetically and have found wide spread use as research tools for gene knockdown. They may also have therapeutic applications.

Silent mutations are mutations in DNA that do not have an observable effect on the organism's phenotype. They are a specific type of neutral mutation. The phrase silent mutation is often used interchangeably with the phrase synonymous mutation; however, synonymous mutations are not always silent, nor vice versa. Synonymous mutations can affect transcription, splicing, mRNA transport, and translation, any of which could alter phenotype, rendering the synonymous mutation non-silent. The substrate specificity of the tRNA to the rare codon can affect the timing of translation, and in turn the co-translational folding of the protein. This is reflected in the codon usage bias that is observed in many species. Mutations that cause the altered codon to produce an amino acid with similar functionality are often classified as silent; if the properties of the amino acid are conserved, this mutation does not usually significantly affect protein function.
Triple-stranded DNA is a DNA structure in which three oligonucleotides wind around each other and form a triple helix. In triple-stranded DNA, the third strand binds to a B-form DNA double helix by forming Hoogsteen base pairs or reversed Hoogsteen hydrogen bonds.
In molecular biology and genetics, the sense of a nucleic acid molecule, particularly of a strand of DNA or RNA, refers to the nature of the roles of the strand and its complement in specifying a sequence of amino acids. Depending on the context, sense may have slightly different meanings. For example, negative-sense strand of DNA is equivalent to the template strand, whereas the positive-sense strand is the non-template strand whose nucleotide sequence is equivalent to the sequence of the mRNA transcript.
Therapeutic gene modulation refers to the practice of altering the expression of a gene at one of various stages, with a view to alleviate some form of ailment. It differs from gene therapy in that gene modulation seeks to alter the expression of an endogenous gene whereas gene therapy concerns the introduction of a gene whose product aids the recipient directly.
Cell-penetrating peptides (CPPs) are short peptides that facilitate cellular intake and uptake of molecules ranging from nanosize particles to small chemical compounds to large fragments of DNA. The "cargo" is associated with the peptides either through chemical linkage via covalent bonds or through non-covalent interactions.
Oblimersen is an antisense oligodeoxyribonucleotide being studied as a possible treatment for several types of cancer, including chronic lymphocytic leukemia, B-cell lymphoma, and breast cancer. It may kill cancer cells by blocking the production of Bcl-2—a protein that makes cancer cells live longer—and by making them more sensitive to chemotherapy.
Natural computing, also called natural computation, is a terminology introduced to encompass three classes of methods: 1) those that take inspiration from nature for the development of novel problem-solving techniques; 2) those that are based on the use of computers to synthesize natural phenomena; and 3) those that employ natural materials to compute. The main fields of research that compose these three branches are artificial neural networks, evolutionary algorithms, swarm intelligence, artificial immune systems, fractal geometry, artificial life, DNA computing, and quantum computing, among others.
In molecular biology, exon skipping is a form of RNA splicing used to cause cells to “skip” over faulty or misaligned sections (exons) of genetic code, leading to a truncated but still functional protein despite the genetic mutation.
Julianna Lisziewicz is a Hungarian immunologist. Lisziewicz headed many research teams that have discovered and produced immunotheraputic drugs to treat diseases like cancer and chronic infections like HIV/AIDS. Some of these drugs have been successfully used in clinical trials.
RNA-targeting small molecules represent a class of small molecules, organic compounds with traditional drug properties that can bind to RNA secondary or tertiary structures and alter translation patterns, localization, and degradation.











