Related Research Articles

Coenzyme A (CoA, SHCoA, CoASH) is a coenzyme, notable for its role in the synthesis and oxidation of fatty acids, and the oxidation of pyruvate in the citric acid cycle. All genomes sequenced to date encode enzymes that use coenzyme A as a substrate, and around 4% of cellular enzymes use it (or a thioester) as a substrate. In humans, CoA biosynthesis requires cysteine, pantothenate (vitamin B5), and adenosine triphosphate (ATP).
Enoyl-CoA-(∆) isomerase (EC 5.3.3.8, also known as dodecenoyl-CoA- isomerase, 3,2-trans-enoyl-CoA isomerase, ∆3 ,∆2 -enoyl-CoA isomerase, or acetylene-allene isomerase, is an enzyme that catalyzes the conversion of cis- or trans-double bonds of coenzyme A bound fatty acids at gamma-carbon to trans double bonds at beta-carbon as below:
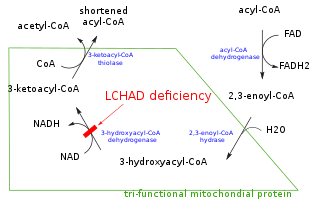
In biochemistry and metabolism, beta-oxidation is the catabolic process by which fatty acid molecules are broken down in the cytosol in prokaryotes and in the mitochondria in eukaryotes to generate acetyl-CoA, which enters the citric acid cycle, and NADH and FADH2, which are co-enzymes used in the electron transport chain. It is named as such because the beta carbon of the fatty acid undergoes oxidation to a carbonyl group. Beta-oxidation is primarily facilitated by the mitochondrial trifunctional protein, an enzyme complex associated with the inner mitochondrial membrane, although very long chain fatty acids are oxidized in peroxisomes.
Acyl-CoA dehydrogenases (ACADs) are a class of enzymes that function to catalyze the initial step in each cycle of fatty acid β-oxidation in the mitochondria of cells. Their action results in the introduction of a trans double-bond between C2 (α) and C3 (β) of the acyl-CoA thioester substrate. Flavin adenine dinucleotide (FAD) is a required co-factor in addition to the presence of an active site glutamate in order for the enzyme to function.

Enoyl-CoA hydratase (ECH) or crotonase is an enzyme EC 4.2.1.17 that hydrates the double bond between the second and third carbons on 2-trans/cis-enoyl-CoA:
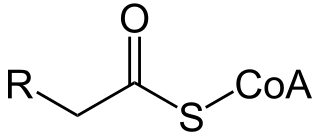
Acyl-CoA is a group of coenzymes that metabolize fatty acids. Acyl-CoA's are susceptible to beta oxidation, forming, ultimately, acetyl-CoA. The acetyl-CoA enters the citric acid cycle, eventually forming several equivalents of ATP. In this way, fats are converted to ATP, the universal biochemical energy carrier.
The long chain fatty acyl-CoA ligase is an enzyme of the ligase family that activates the oxidation of complex fatty acids. Long chain fatty acyl-CoA synthetase catalyzes the formation of fatty acyl-CoA by a two-step process proceeding through an adenylated intermediate. The enzyme catalyzes the following reaction,

2,4 Dienoyl-CoA reductase also known as DECR1 is an enzyme which in humans is encoded by the DECR1 gene which resides on chromosome 8. This enzyme catalyzes the following reactions

Thiolases, also known as acetyl-coenzyme A acetyltransferases (ACAT), are enzymes which convert two units of acetyl-CoA to acetoacetyl CoA in the mevalonate pathway.

3-Methylglutaconyl-CoA hydratase, also known as MG-CoA hydratase and AUH, is an enzyme encoded by the AUH gene on chromosome 19. It is a member of the enoyl-CoA hydratase/isomerase superfamily, but it is the only member of that family that is able to bind to RNA. Not only does it bind to RNA, AUH has also been observed to be involved in the metabolic enzymatic activity, making it a dual-role protein. Mutations of this gene have been found to cause a disease called 3-Methylglutaconic Acuduria Type 1.

In enzymology, an acyl-CoA dehydrogenase (NADP+) (EC 1.3.1.8) is an enzyme that catalyzes the chemical reaction
In enzymology, a trans-2-decenoyl-[acyl-carrier protein] isomerase is an enzyme that catalyzes the chemical reaction
In enzymology, a vinylacetyl-CoA Delta-isomerase is an enzyme that catalyzes the chemical reaction
In enzymology, a crotonoyl-[acyl-carrier-protein] hydratase (EC 4.2.1.58) is an enzyme that catalyzes the chemical reaction
The enzyme long-chain-enoyl-CoA hydratase (EC 4.2.1.74) catalyzes the chemical reaction

Enoyl Coenzyme A hydratase, short chain, 1, mitochondrial, also known as ECHS1, is a human gene.

Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial is an enzyme that in humans is encoded by the ECH1 gene.
3-hydroxydecanoyl-(acyl-carrier-protein) dehydratase (EC 4.2.1.60, D-3-hydroxydecanoyl-[acyl-carrier protein] dehydratase, 3-hydroxydecanoyl-acyl carrier protein dehydrase, 3-hydroxydecanoyl-acyl carrier protein dehydratase, β-hydroxydecanoyl thioester dehydrase, β-hydroxydecanoate dehydrase, beta-hydroxydecanoyl thiol ester dehydrase, FabA, β-hydroxyacyl-acyl carrier protein dehydratase, HDDase, β-hydroxyacyl-ACP dehydrase, (3R)-3-hydroxydecanoyl-[acyl-carrier-protein] hydro-lyase) is an enzyme with systematic name (3R)-3-hydroxydecanoyl-(acyl-carrier protein) hydro-lyase. This enzyme catalyses the following chemical reaction
Enoyl-CoA hydratase 2 is an enzyme with systematic name (3R)-3-hydroxyacyl-CoA hydro-lyase. This enzyme catalyses the following chemical reaction on D-3-hydroxyacyl-CoA

Ketoacyl synthases (KSs) catalyze the condensation reaction of acyl-CoA or acyl-acyl ACP with malonyl-CoA to form 3-ketoacyl-CoA or with malonyl-ACP to form 3-ketoacyl-ACP. This reaction is a key step in the fatty acid synthesis cycle, as the resulting acyl chain is two carbon atoms longer than before. KSs exist as individual enzymes, as they do in type II fatty acid synthesis and type II polyketide synthesis, or as domains in large multidomain enzymes, such as type I fatty acid synthases (FASs) and polyketide synthases (PKSs). KSs are divided into five families: KS1, KS2, KS3, KS4, and KS5.
References
- ↑ Gerlt JA, Benning MM, Holden HM, Haller T (2001). "The crotonase superfamily: divergently related enzymes that catalyze different reactions involving acyl coenzyme a thioesters". Acc. Chem. Res. 34 (2): 145–57. doi:10.1021/ar000053l. PMID 11263873.
- ↑ Brzozowski AM, Leonard PM, Bennett JP, Whittingham JL, Grogan G (2007). "Structural characterization of a beta-diketone hydrolase from the cyanobacterium Anabaena sp. PCC 7120 in native and product-bound forms, a coenzyme A-independent member of the crotonase suprafamily". Biochemistry. 46 (1): 137–44. doi:10.1021/bi061900g. PMID 17198383.
- ↑ Wu J, Kisker C, Whitty A, Feng Y, Rudolph MJ, Bell AF, Hofstein HA, Parikh S, Tonge PJ (2002). "Stereoselectivity of enoyl-CoA hydratase results from preferential activation of one of two bound substrate conformers". Chem. Biol. 9 (11): 1247–55. doi: 10.1016/S1074-5521(02)00263-6 . PMID 12445775.
- ↑ Stoffel W, Muller-Newen G (1991). "Mitochondrial 3-2trans-Enoyl-CoA isomerase. Purification, cloning, expression, and mitochondrial import of the key enzyme of unsaturated fatty acid beta-oxidation". Biol. Chem. Hoppe-Seyler. 372 (8): 613–624. doi:10.1515/bchm3.1991.372.2.613. PMID 1958319.
- ↑ Dunaway-Mariano D, Benning MM, Wesenberg G, Holden HM, Taylor KL, Yang G, Liu R-Q, Xiang H (1996). "Structure of 4-chlorobenzoyl coenzyme A dehalogenase determined to 1.8 A resolution: an enzyme catalyst generated via adaptive mutation". Biochemistry. 35 (25): 8103–9. doi:10.1021/bi960768p. PMID 8679561.
- ↑ Hiltunen JK, Wierenga RK, Modis Y, Filppula SA, Novikov DK, Norledge B (1998). "The crystal structure of dienoyl-CoA isomerase at 1.5 A resolution reveals the importance of aspartate and glutamate sidechains for catalysis". Structure. 6 (8): 957–70. doi: 10.1016/s0969-2126(98)00098-7 . PMID 9739087.
- ↑ Baker EN, Johnston JM, Arcus VL (2005). "Structure of naphthoate synthase (MenB) from Mycobacterium tuberculosis in both native and product-bound forms". Acta Crystallogr. D. 61 (Pt 9): 1199–206. doi:10.1107/S0907444905017531. PMID 16131752.
- ↑ Kleber HP, Elssner T, Engemann C, Baumgart K (2001). "Involvement of coenzyme A esters and two new enzymes, an enoyl-CoA hydratase and a CoA-transferase, in the hydration of crotonobetaine to L-carnitine by Escherichia coli". Biochemistry. 40 (37): 11140–8. doi:10.1021/bi0108812. PMID 11551212.
- ↑ Gerlt JA, Benning MM, Holden HM, Haller T (2000). "New reactions in the crotonase superfamily: structure of methylmalonyl CoA decarboxylase from Escherichia coli". Biochemistry. 39 (16): 4630–9. doi:10.1021/bi9928896. PMID 10769118.
- ↑ Schofield CJ, McDonough MA, Sleeman MC, Sorensen JL, Batchelar ET (2005). "Structural and mechanistic studies on carboxymethylproline synthase (CarB), a unique member of the crotonase superfamily catalyzing the first step in carbapenem biosynthesis". J. Biol. Chem. 280 (41): 34956–65. doi: 10.1074/jbc.M507196200 . PMID 16096274.
- ↑ Leonard PM, Grogan G (2004). "Structure of 6-oxo camphor hydrolase H122A mutant bound to its natural product, (2S,4S)-alpha-campholinic acid: mutant structure suggests an atypical mode of transition state binding for a crotonase homolog". J. Biol. Chem. 279 (30): 31312–17. doi: 10.1074/jbc.M403514200 . PMID 15138275.
- ↑ Resibois-Gregoire A, Dourov N (1966). "Electron microscopic study of a case of cerebral glycogenosis". Acta Neuropathol. 6 (1): 70–9. doi:10.1007/BF00691083. PMID 5229654. S2CID 21331079.
- ↑ Nureki O, Fukai S, Yokoyama S, Muto Y, Kurimoto K (2001). "Crystal structure of human AUH protein, a single-stranded RNA binding homolog of enoyl-CoA hydratase". Structure. 9 (12): 1253–63. doi: 10.1016/S0969-2126(01)00686-4 . PMID 11738050.