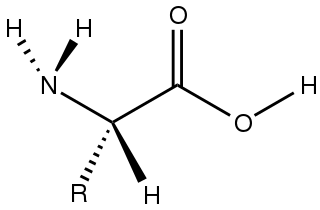
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins. Only these 22 appear in the genetic code of life.
The urea cycle (also known as the ornithine cycle) is a cycle of biochemical reactions that produces urea (NH2)2CO from ammonia (NH3). Animals that use this cycle, mainly amphibians and mammals, are called ureotelic.

Arginine is the amino acid with the formula (H2N)(HN)CN(H)(CH2)3CH(NH2)CO2H. The molecule features a guanidino group appended to a standard amino acid framework. At physiological pH, the carboxylic acid is deprotonated (−CO2−) and both the amino and guanidino groups are protonated, resulting in a cation. Only the l-arginine (symbol Arg or R) enantiomer is found naturally. Arg residues are common components of proteins. It is encoded by the codons CGU, CGC, CGA, CGG, AGA, and AGG. The guanidine group in arginine is the precursor for the biosynthesis of nitric oxide. Like all amino acids, it is a white, water-soluble solid.
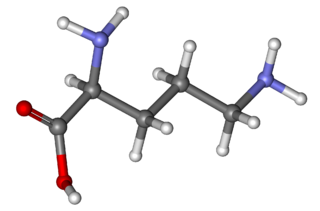
Ornithine is a non-proteinogenic α-amino acid that plays a role in the urea cycle. Ornithine is abnormally accumulated in the body in ornithine transcarbamylase deficiency. The radical is ornithyl.

Carbamoyl phosphate is an anion of biochemical significance. In land-dwelling animals, it is an intermediary metabolite in nitrogen disposal through the urea cycle and the synthesis of pyrimidines. Its enzymatic counterpart, carbamoyl phosphate synthetase I, interacts with a class of molecules called sirtuins, NAD dependent protein deacetylases, and ATP to form carbamoyl phosphate. CP then enters the urea cycle in which it reacts with ornithine to form citrulline.
In molecular biology, biosynthesis is a multi-step, enzyme-catalyzed process where substrates are converted into more complex products in living organisms. In biosynthesis, simple compounds are modified, converted into other compounds, or joined to form macromolecules. This process often consists of metabolic pathways. Some of these biosynthetic pathways are located within a single cellular organelle, while others involve enzymes that are located within multiple cellular organelles. Examples of these biosynthetic pathways include the production of lipid membrane components and nucleotides. Biosynthesis is usually synonymous with anabolism.

In the mitochondrion, the matrix is the space within the inner membrane. The word "matrix" stems from the fact that this space is viscous, compared to the relatively aqueous cytoplasm. The mitochondrial matrix contains the mitochondrial DNA, ribosomes, soluble enzymes, small organic molecules, nucleotide cofactors, and inorganic ions.[1] The enzymes in the matrix facilitate reactions responsible for the production of ATP, such as the citric acid cycle, oxidative phosphorylation, oxidation of pyruvate, and the beta oxidation of fatty acids.

Argininosuccinate synthase or synthetase is an enzyme that catalyzes the synthesis of argininosuccinate from citrulline and aspartate. In humans, argininosuccinate synthase is encoded by the ASS gene located on chromosome 9.

The enzyme argininosuccinate lyase (EC 4.3.2.1, ASL, argininosuccinase; systematic name 2-(N ω-L-arginino)succinate arginine-lyase (fumarate-forming)) catalyzes the reversible breakdown of argininosuccinate:
Phytotoxins are substances that are poisonous or toxic to the growth of plants. Phytotoxic substances may result from human activity, as with herbicides, or they may be produced by plants, by microorganisms, or by naturally occurring chemical reactions.

Amino acid synthesis is the set of biochemical processes by which the amino acids are produced. The substrates for these processes are various compounds in the organism's diet or growth media. Not all organisms are able to synthesize all amino acids. For example, humans can synthesize 11 of the 20 standard amino acids. These 11 are called the non-essential amino acids).

The enzyme Acid-Induced Arginine Decarboxylase (AdiA), also commonly referred to as arginine decarboxylase, catalyzes the conversion of L-arginine into agmatine and carbon dioxide. The process consumes a proton in the decarboxylation and employs a pyridoxal-5'-phosphate (PLP) cofactor, similar to other enzymes involved in amino acid metabolism, such as ornithine decarboxylase and glutamine decarboxylase. It is found in bacteria and virus, though most research has so far focused on forms of the enzyme in bacteria. During the AdiA catalyzed decarboxylation of arginine, the necessary proton is consumed from the cell cytoplasm which helps to prevent the over-accumulation of protons inside the cell and serves to increase the intracellular pH. Arginine decarboxylase is part of an enzymatic system in Escherichia coli, Salmonella Typhimurium, and methane-producing bacteria Methanococcus jannaschii that makes these organisms acid resistant and allows them to survive under highly acidic medium.

In enzymology, an arginine—tRNA ligase is an enzyme that catalyzes the chemical reaction
In enzymology, an arginine deiminase (EC 3.5.3.6) is an enzyme that catalyzes the chemical reaction
Delta-1-pyrroline-5-carboxylate synthetase (P5CS) is an enzyme that in humans is encoded by the ALDH18A1 gene. This gene is a member of the aldehyde dehydrogenase family and encodes a bifunctional ATP- and NADPH-dependent mitochondrial enzyme with both gamma-glutamyl kinase and gamma-glutamyl phosphate reductase activities. The encoded protein catalyzes the reduction of glutamate to delta1-pyrroline-5-carboxylate, a critical step in the de novo biosynthesis of proline, ornithine and arginine. Mutations in this gene lead to hyperammonemia, hypoornithinemia, hypocitrullinemia, hypoargininemia and hypoprolinemia and may be associated with neurodegeneration, cataracts and connective tissue diseases. Alternatively spliced transcript variants, encoding different isoforms, have been described for this gene. As reported by Bruno Reversade and colleagues, ALDH18A1 deficiency or dominant-negative mutations in P5CS in humans causes a progeroid disease known as De Barsy Syndrome.

l-Canaline ) is a non-proteinogenic amino acid. The compound is found in legumes that contain canavanine, from which it is produced by the action of arginase. The most common-used source for this amino acid is the jack bean, Canavalia ensiformis.
The bacterial cell wall provides strength and rigidity to counteract internal osmotic pressure, and protection against the environment. The peptidoglycan layer gives the cell wall its strength, and helps maintain the overall shape of the cell. The basic peptidoglycan structure of both Gram-positive and Gram-negative bacteria comprises a sheet of glycan chains connected by short cross-linking polypeptides. Biosynthesis of peptidoglycan is a multi-step process comprising three main stages:
- formation of UDP-N-acetylmuramic acid (UDPMurNAc) from N-acetylglucosamine (GlcNAc).
- addition of a short polypeptide chain to the UDPMurNAc.
- addition of a second GlcNAc to the disaccharide-pentapeptide building block and transport of this unit through the cytoplasmic membrane and incorporation into the growing peptidoglycan layer.

Cyanophycinase (EC 3.4.15.6, cyanophycin degrading enzyme, beta-Asp-Arg hydrolysing enzyme, CGPase, CphB, CphE, cyanophycin granule polypeptidase, extracellular CGPase) is an enzyme. It catalyses the following chemical reaction
Cyanophycin synthase (L-aspartate-adding) is an enzyme with systematic name cyanophycin:L-aspartate ligase (ADP-forming). This enzyme catalyses the following chemical reaction
Cyanophycin synthase (L-arginine-adding) is an enzyme with systematic name cyanophycin:L-arginine ligase (ADP-forming). This enzyme catalyses the following chemical reaction:













