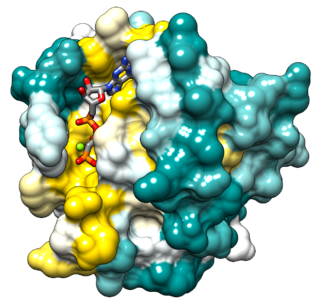
Ras, from "Rat sarcoma virus", is a family of related proteins that are expressed in all animal cell lineages and organs. All Ras protein family members belong to a class of protein called small GTPase, and are involved in transmitting signals within cells. Ras is the prototypical member of the Ras superfamily of proteins, which are all related in three-dimensional structure and regulate diverse cell behaviours.
GTPase-activating proteins or GTPase-accelerating proteins (GAPs) are a family of regulatory proteins whose members can bind to activated G proteins and stimulate their GTPase activity, with the result of terminating the signaling event. GAPs are also known as RGS protein, or RGS proteins, and these proteins are crucial in controlling the activity of G proteins. Regulation of G proteins is important because these proteins are involved in a variety of important cellular processes. The large G proteins, for example, are involved in transduction of signaling from the G protein-coupled receptor for a variety of signaling processes like hormonal signaling, and small G proteins are involved in processes like cellular trafficking and cell cycling. GAP's role in this function is to turn the G protein's activity off. In this sense, GAPs function is opposite to that of guanine nucleotide exchange factors (GEFs), which serve to enhance G protein signaling.

The KRAS gene provides instructions for making a protein called K-Ras, part of the RAS/MAPK pathway. The protein relays signals from outside the cell to the cell's nucleus. These signals instruct the cell to grow and divide (proliferate) or to mature and take on specialized functions (differentiate). The K-Ras protein is a GTPase, which means it converts a molecule called GTP into another molecule called GDP. In this way the K-Ras protein acts like a switch that is turned on and off by the GTP and GDP molecules. To transmit signals, it must be turned on by attaching (binding) to a molecule of GTP. The K-Ras protein is turned off (inactivated) when it converts the GTP to GDP. When the protein is bound to GDP, it does not relay signals to the cell's nucleus. It is called KRAS because it was first identified as an oncogene in KirstenRAt Sarcoma virus. The viral oncogene was derived from cellular genome. Thus, KRAS gene in cellular genome is called a proto-oncogene.

Cyclin D1 is a protein that in humans is encoded by the CCND1 gene.

Transforming protein RhoA, also known as Ras homolog family member A (RhoA), is a small GTPase protein in the Rho family of GTPases that in humans is encoded by the RHOA gene. While the effects of RhoA activity are not all well known, it is primarily associated with cytoskeleton regulation, mostly actin stress fibers formation and actomyosin contractility. It acts upon several effectors. Among them, ROCK1 and DIAPH1 are the best described. RhoA, and the other Rho GTPases, are part of a larger family of related proteins known as the Ras superfamily, a family of proteins involved in the regulation and timing of cell division. RhoA is one of the oldest Rho GTPases, with homologues present in the genomes since 1.5 billion years. As a consequence, RhoA is somehow involved in many cellular processes which emerged throughout evolution. RhoA specifically is regarded as a prominent regulatory factor in other functions such as the regulation of cytoskeletal dynamics, transcription, cell cycle progression and cell transformation.

Ras association domain-containing protein 1 is a protein that in humans is encoded by the RASSF1 gene.
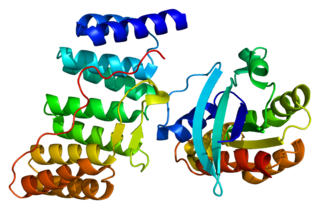
Ras-related C3 botulinum toxin substrate 3 (Rac3) is a G protein that in humans is encoded by the RAC3 gene. It is an important component of intracellular signalling pathways. Rac3 is a member of the Rac subfamily of the Rho family of small G proteins. Members of this superfamily appear to regulate a diverse array of cellular events, including the control of cell growth, cytoskeletal reorganization, and the activation of protein kinases.

RhoC is a small signaling G protein, and is a member of the Rac subfamily of the family Rho family of GTPases. It is encoded by the gene RHOC.

Ras association domain-containing protein 5 is a protein that in humans is encoded by the RASSF5 or F5 gene.

H19 is a gene for a long noncoding RNA, found in humans and elsewhere. H19 has a role in the negative regulation of body weight and cell proliferation. This gene also has a role in the formation of some cancers and in the regulation of gene expression. .

Deleted in Liver Cancer 1 also known as DLC1 and StAR-related lipid transfer protein 12 (STARD12) is a protein which in humans is encoded by the DLC1 gene.

Rho GDP-dissociation inhibitor 2 is a protein that in humans is encoded by the ARHGDIB gene. Aliases of this gene include RhoGDI2, GDID4, Rho GDI 2, and others.

Large tumor suppressor kinase 1 (LATS1) is an enzyme that in humans is encoded by the LATS1 gene.

ID4 is a protein coding gene. In humans, it encodes for the protein known as DNA-binding protein inhibitor ID-4. This protein is known to be involved in the regulation of many cellular processes during both prenatal development and tumorigenesis. This is inclusive of embryonic cellular growth, senescence, cellular differentiation, apoptosis, and as an oncogene in angiogenesis.

CDKN2A, also known as cyclin-dependent kinase inhibitor 2A, is a gene which in humans is located at chromosome 9, band p21.3. It is ubiquitously expressed in many tissues and cell types. The gene codes for two proteins, including the INK4 family member p16 and p14arf. Both act as tumor suppressors by regulating the cell cycle. p16 inhibits cyclin dependent kinases 4 and 6 and thereby activates the retinoblastoma (Rb) family of proteins, which block traversal from G1 to S-phase. p14ARF activates the p53 tumor suppressor. Somatic mutations of CDKN2A are common in the majority of human cancers, with estimates that CDKN2A is the second most commonly inactivated gene in cancer after p53. Germline mutations of CDKN2A are associated with familial melanoma, glioblastoma and pancreatic cancer. The CDKN2A gene also contains one of 27 SNPs associated with increased risk of coronary artery disease.
A metastasis suppressor is a protein that acts to slow or prevent metastases from spreading in the body of an organism with cancer. Metastasis is one of the most lethal cancer processes. This process is responsible for about ninety percent of human cancer deaths. Proteins that act to slow or prevent metastases are different from those that act to suppress tumor growth. Genes for about a dozen such proteins are known in humans and other animals.
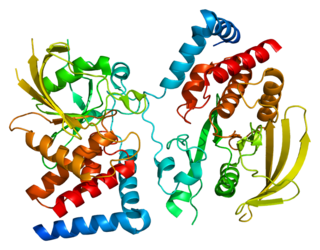
Receptor-type tyrosine-protein phosphatase gamma is an enzyme that in humans is encoded by the PTPRG gene.

Epithelial membrane protein 3 (EMP3) is a trans-membrane signaling molecule that is encoded by the myelin-related gene EMP3. EMP3 is a member of the peripheral myelin protein gene family 22-kDa (PMP22), which is mainly responsible for the formation of the sheath of compact myelin. Although the detailed functions and mechanisms of EMP3 still remain unclear, it is suggested that EMP3 is possibly epigenetically linked to certain carcinomas.

In molecular biology, miR-137 is a short non-coding RNA molecule that functions to regulate the expression levels of other genes by various mechanisms. miR-137 is located on human chromosome 1p22 and has been implicated to act as a tumor suppressor in several cancer types including colorectal cancer, squamous cell carcinoma and melanoma via cell cycle control.

Augurin is a protein that in humans is encoded by the C2orf40 gene.


















