
The cell cycle, or cell-division cycle, is the series of events that take place in a cell that causes it to divide into two daughter cells. These events include the duplication of its DNA and some of its organelles, and subsequently the partitioning of its cytoplasm, chromosomes and other components into two daughter cells in a process called cell division.

In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part of biological inheritance. This is essential for cell division during growth and repair of damaged tissues, while it also ensures that each of the new cells receives its own copy of the DNA. The cell possesses the distinctive property of division, which makes replication of DNA essential.

The origin of replication is a particular sequence in a genome at which replication is initiated. Propagation of the genetic material between generations requires timely and accurate duplication of DNA by semiconservative replication prior to cell division to ensure each daughter cell receives the full complement of chromosomes. This can either involve the replication of DNA in living organisms such as prokaryotes and eukaryotes, or that of DNA or RNA in viruses, such as double-stranded RNA viruses. Synthesis of daughter strands starts at discrete sites, termed replication origins, and proceeds in a bidirectional manner until all genomic DNA is replicated. Despite the fundamental nature of these events, organisms have evolved surprisingly divergent strategies that control replication onset. Although the specific replication origin organization structure and recognition varies from species to species, some common characteristics are shared.
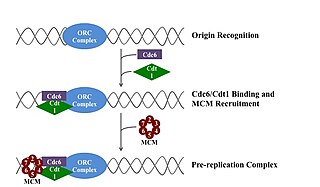
A pre-replication complex (pre-RC) is a protein complex that forms at the origin of replication during the initiation step of DNA replication. Formation of the pre-RC is required for DNA replication to occur. Complete and faithful replication of the genome ensures that each daughter cell will carry the same genetic information as the parent cell. Accordingly, formation of the pre-RC is a very important part of the cell cycle.

G2 phase, Gap 2 phase, or Growth 2 phase, is the third subphase of interphase in the cell cycle directly preceding mitosis. It follows the successful completion of S phase, during which the cell’s DNA is replicated. G2 phase ends with the onset of prophase, the first phase of mitosis in which the cell’s chromatin condenses into chromosomes.
Endoreduplication is replication of the nuclear genome in the absence of mitosis, which leads to elevated nuclear gene content and polyploidy. Endoreduplication can be understood simply as a variant form of the mitotic cell cycle (G1-S-G2-M) in which mitosis is circumvented entirely, due to modulation of cyclin-dependent kinase (CDK) activity. Examples of endoreduplication characterised in arthropod, mammalian, and plant species suggest that it is a universal developmental mechanism responsible for the differentiation and morphogenesis of cell types that fulfill an array of biological functions. While endoreduplication is often limited to specific cell types in animals, it is considerably more widespread in plants, such that polyploidy can be detected in the majority of plant tissues. Polyploidy and aneuploidy are common phenomena in cancer cells. Given that oncogenesis and endoreduplication likely involve subversion of common cell cycle regulatory mechanisms, a thorough understanding of endoreduplication may provide important insights for cancer biology.
A licensing factor is a protein or complex of proteins that allows an origin of replication to begin DNA replication at that site. Licensing factors primarily occur in eukaryotic cells, since bacteria use simpler systems to initiate replication. However, many archaea use homologues of eukaryotic licensing factors to initiate replication.
Cyclin A is a member of the cyclin family, a group of proteins that function in regulating progression through the cell cycle. The stages that a cell passes through that culminate in its division and replication are collectively known as the cell cycle Since the successful division and replication of a cell is essential for its survival, the cell cycle is tightly regulated by several components to ensure the efficient and error-free progression through the cell cycle. One such regulatory component is cyclin A which plays a role in the regulation of two different cell cycle stages.

Geminin, DNA replication inhibitor, also known as GMNN, is a protein in humans encoded by the GMNN gene. A nuclear protein present in most eukaryotes and highly conserved across species, numerous functions have been elucidated for geminin including roles in metazoan cell cycle, cellular proliferation, cell lineage commitment, and neural differentiation. One example of its function is the inhibition of Cdt1.
In molecular biology, origin recognition complex (ORC) is a multi-subunit DNA binding complex that binds in all eukaryotes and archaea in an ATP-dependent manner to origins of replication. The subunits of this complex are encoded by the ORC1, ORC2, ORC3, ORC4, ORC5 and ORC6 genes. ORC is a central component for eukaryotic DNA replication, and remains bound to chromatin at replication origins throughout the cell cycle.

Eukaryotic DNA replication is a conserved mechanism that restricts DNA replication to once per cell cycle. Eukaryotic DNA replication of chromosomal DNA is central for the duplication of a cell and is necessary for the maintenance of the eukaryotic genome.

The minichromosome maintenance protein complex (MCM) is a DNA helicase essential for genomic DNA replication. Eukaryotic MCM consists of six gene products, Mcm2–7, which form a heterohexamer. As a critical protein for cell division, MCM is also the target of various checkpoint pathways, such as the S-phase entry and S-phase arrest checkpoints. Both the loading and activation of MCM helicase are strictly regulated and are coupled to cell growth cycles. Deregulation of MCM function has been linked to genomic instability and a variety of carcinomas.

DNA replication licensing factor MCM2 is a protein that in humans is encoded by the MCM2 gene.

CDT1 is a protein that in humans is encoded by the CDT1 gene. It is a licensing factor that functions to limit DNA from replicating more than once per cell cycle.

Cell division cycle 7-related protein kinase is an enzyme that in humans is encoded by the CDC7 gene. The Cdc7 kinase is involved in regulation of the cell cycle at the point of chromosomal DNA replication. The gene CDC7 appears to be conserved throughout eukaryotic evolution; this means that most eukaryotic cells have the Cdc7 kinase protein.

Cdc6, or cell division cycle 6, is a protein in eukaryotic cells. It is mainly studied in the budding yeast Saccharomyces cerevisiae. It is an essential regulator of DNA replication and plays important roles in the activation and maintenance of the checkpoint mechanisms in the cell cycle that coordinate S phase and mitosis. It is part of the pre-replicative complex (pre-RC) and is required for loading minichromosome maintenance (MCM) proteins onto the DNA, an essential step in the initiation of DNA synthesis. In addition, it is a member of the family of AAA+ ATPases and highly related to ORC1; both are the same protein in archaea.

In cell biology, eukaryotes possess a regulatory system that ensures that DNA replication occurs only once per cell cycle.

Origin recognition complex subunit 1 is a protein that in humans is encoded by the ORC1 gene. It is closely related to CDC6, and both are the same protein in archaea.
Anindya Dutta is an Indian-born American biochemist and cancer researcher, a Chair of the Department of Genetics at the University of Alabama at Birmingham School of Medicine since 2021, who has served as Chair of the Department of Biochemistry and Molecular Genetics at the University of Virginia School of Medicine in 2011–2021. Dutta's research has focused on the mammalian cell cycle with an emphasis on DNA replication and repair and on noncoding RNAs. He is particularly interested in how de-regulation of these processes promote cancer progression. For his accomplishments he has been elected a Fellow of the American Association for the Advancement of Science, received the Ranbaxy Award in Biomedical Sciences, the Outstanding Investigator Award from the American Society for Investigative Pathology, the Distinguished Scientist Award from the University of Virginia and the Mark Brothers Award from the Indiana University School of Medicine.
Julian Blow is a molecular biologist, Professor of Chromosome Maintenance, and also the Dean of the School of Life Sciences, University of Dundee, Scotland.















