Related Research Articles

Supervised learning (SL) is a paradigm in machine learning where input objects and a desired output value train a model. The training data is processed, building a function that maps new data on expected output values. An optimal scenario will allow for the algorithm to correctly determine output values for unseen instances. This requires the learning algorithm to generalize from the training data to unseen situations in a "reasonable" way. This statistical quality of an algorithm is measured through the so-called generalization error.

In computer science and operations research, a genetic algorithm (GA) is a metaheuristic inspired by the process of natural selection that belongs to the larger class of evolutionary algorithms (EA). Genetic algorithms are commonly used to generate high-quality solutions to optimization and search problems by relying on biologically inspired operators such as mutation, crossover and selection. Some examples of GA applications include optimizing decision trees for better performance, solving sudoku puzzles, hyperparameter optimization, causal inference, etc.
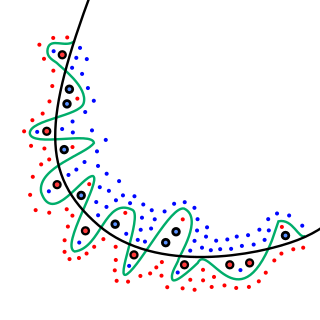
In mathematical modeling, overfitting is "the production of an analysis that corresponds too closely or exactly to a particular set of data, and may therefore fail to fit to additional data or predict future observations reliably". An overfitted model is a mathematical model that contains more parameters than can be justified by the data. In a mathematical sense, these parameters represent the degree of a polynomial. The essence of overfitting is to have unknowingly extracted some of the residual variation as if that variation represented underlying model structure.
Machine learning (ML) is a field of study in artificial intelligence concerned with the development and study of statistical algorithms that can learn from data and generalize to unseen data and thus perform tasks without explicit instructions. Recently, artificial neural networks have been able to surpass many previous approaches in performance.

In computer science, evolutionary computation is a family of algorithms for global optimization inspired by biological evolution, and the subfield of artificial intelligence and soft computing studying these algorithms. In technical terms, they are a family of population-based trial and error problem solvers with a metaheuristic or stochastic optimization character.

Cross-validation, sometimes called rotation estimation or out-of-sample testing, is any of various similar model validation techniques for assessing how the results of a statistical analysis will generalize to an independent data set. Cross-validation includes resampling and sample splitting methods that use different portions of the data to test and train a model on different iterations. It is often used in settings where the goal is prediction, and one wants to estimate how accurately a predictive model will perform in practice. It can also be used to assess the quality of a fitted model and the stability of its parameters.
Association rule learning is a rule-based machine learning method for discovering interesting relations between variables in large databases. It is intended to identify strong rules discovered in databases using some measures of interestingness. In any given transaction with a variety of items, association rules are meant to discover the rules that determine how or why certain items are connected.

Learning classifier systems, or LCS, are a paradigm of rule-based machine learning methods that combine a discovery component with a learning component. Learning classifier systems seek to identify a set of context-dependent rules that collectively store and apply knowledge in a piecewise manner in order to make predictions. This approach allows complex solution spaces to be broken up into smaller, simpler parts for the reinforcement learning that is inside artificial intelligence research.
Random sample consensus (RANSAC) is an iterative method to estimate parameters of a mathematical model from a set of observed data that contains outliers, when outliers are to be accorded no influence on the values of the estimates. Therefore, it also can be interpreted as an outlier detection method. It is a non-deterministic algorithm in the sense that it produces a reasonable result only with a certain probability, with this probability increasing as more iterations are allowed. The algorithm was first published by Fischler and Bolles at SRI International in 1981. They used RANSAC to solve the Location Determination Problem (LDP), where the goal is to determine the points in the space that project onto an image into a set of landmarks with known locations.
In computer programming, gene expression programming (GEP) is an evolutionary algorithm that creates computer programs or models. These computer programs are complex tree structures that learn and adapt by changing their sizes, shapes, and composition, much like a living organism. And like living organisms, the computer programs of GEP are also encoded in simple linear chromosomes of fixed length. Thus, GEP is a genotype–phenotype system, benefiting from a simple genome to keep and transmit the genetic information and a complex phenotype to explore the environment and adapt to it.
Bootstrap aggregating, also called bagging, is a machine learning ensemble meta-algorithm designed to improve the stability and accuracy of machine learning algorithms used in statistical classification and regression. It also reduces variance and helps to avoid overfitting. Although it is usually applied to decision tree methods, it can be used with any type of method. Bagging is a special case of the model averaging approach.
In machine learning, a common task is the study and construction of algorithms that can learn from and make predictions on data. Such algorithms function by making data-driven predictions or decisions, through building a mathematical model from input data. These input data used to build the model are usually divided into multiple data sets. In particular, three data sets are commonly used in different stages of the creation of the model: training, validation, and test sets.
In machine learning, multi-label classification or multi-output classification is a variant of the classification problem where multiple nonexclusive labels may be assigned to each instance. Multi-label classification is a generalization of multiclass classification, which is the single-label problem of categorizing instances into precisely one of several classes. In the multi-label problem the labels are nonexclusive and there is no constraint on how many of the classes the instance can be assigned to.
In statistics, regression validation is the process of deciding whether the numerical results quantifying hypothesized relationships between variables, obtained from regression analysis, are acceptable as descriptions of the data. The validation process can involve analyzing the goodness of fit of the regression, analyzing whether the regression residuals are random, and checking whether the model's predictive performance deteriorates substantially when applied to data that were not used in model estimation.
Contrast set learning is a form of association rule learning that seeks to identify meaningful differences between separate groups by reverse-engineering the key predictors that identify for each particular group. For example, given a set of attributes for a pool of students, a contrast set learner would identify the contrasting features between students seeking bachelor's degrees and those working toward PhD degrees.

Symbolic regression (SR) is a type of regression analysis that searches the space of mathematical expressions to find the model that best fits a given dataset, both in terms of accuracy and simplicity.
Out-of-bag (OOB) error, also called out-of-bag estimate, is a method of measuring the prediction error of random forests, boosted decision trees, and other machine learning models utilizing bootstrap aggregating (bagging). Bagging uses subsampling with replacement to create training samples for the model to learn from. OOB error is the mean prediction error on each training sample xi, using only the trees that did not have xi in their bootstrap sample.
The following outline is provided as an overview of and topical guide to machine learning:
Machine learning in bioinformatics is the application of machine learning algorithms to bioinformatics, including genomics, proteomics, microarrays, systems biology, evolution, and text mining.
In machine learning, hyperparameter optimization or tuning is the problem of choosing a set of optimal hyperparameters for a learning algorithm. A hyperparameter is a parameter whose value is used to control the learning process.
References
- ↑ Wai-Ho Au, Keith C. C. Chan, and Xin Yao. "A Novel Evolutionary Data Mining Algorithm With Applications to Churn Prediction", IEEE , retrieved on 2008-12-4.
- 1 2 3 4 5 6 7 8 9 10 11 Freitas, Alex A. "A Survey of Evolutionary Algorithms for Data Mining and Knowledge Discovery", Pontifícia Universidade Católica do Paraná , Retrieved on 2008-12-4.
- 1 2 3 4 5 6 7 8 9 10 11 Jiawei Han, Micheline Kamber Data Mining: Concepts and Techniques (2006), Morgan Kaufmann, ISBN 1-55860-901-6