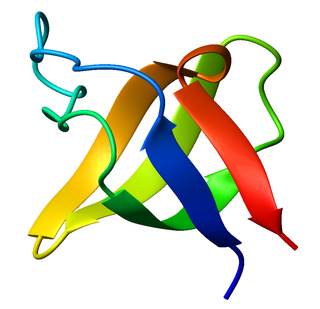
The SRC Homology 3 Domain is a small protein domain of about 60 amino acid residues. Initially, SH3 was described as a conserved sequence in the viral adaptor protein v-Crk. This domain is also present in the molecules of phospholipase and several cytoplasmic tyrosine kinases such as Abl and Src. It has also been identified in several other protein families such as: PI3 Kinase, Ras GTPase-activating protein, CDC24 and cdc25. SH3 domains are found in proteins of signaling pathways regulating the cytoskeleton, the Ras protein, and the Src kinase and many others. The SH3 proteins interact with adaptor proteins and tyrosine kinases. Interacting with tyrosine kinases, SH3 proteins usually bind far away from the active site. Approximately 300 SH3 domains are found in proteins encoded in the human genome. In addition to that, the SH3 domain was responsible for controlling protein-protein interactions in the signal transduction pathways and regulating the interactions of proteins involved in the cytoplasmic signaling.

A leucine zipper is a common three-dimensional structural motif in proteins. They were first described by Landschulz and collaborators in 1988 when they found that an enhancer binding protein had a very characteristic 30-amino acid segment and the display of these amino acid sequences on an idealized alpha helix revealed a periodic repetition of leucine residues at every seventh position over a distance covering eight helical turns. The polypeptide segments containing these periodic arrays of leucine residues were proposed to exist in an alpha-helical conformation and the leucine side chains from one alpha helix interdigitate with those from the alpha helix of a second polypeptide, facilitating dimerization.

The immunoglobulin superfamily (IgSF) is a large protein superfamily of cell surface and soluble proteins that are involved in the recognition, binding, or adhesion processes of cells. Molecules are categorized as members of this superfamily based on shared structural features with immunoglobulins ; they all possess a domain known as an immunoglobulin domain or fold. Members of the IgSF include cell surface antigen receptors, co-receptors and co-stimulatory molecules of the immune system, molecules involved in antigen presentation to lymphocytes, cell adhesion molecules, certain cytokine receptors and intracellular muscle proteins. They are commonly associated with roles in the immune system. Otherwise, the sperm-specific protein IZUMO1, a member of the immunoglobulin superfamily, has also been identified as the only sperm membrane protein essential for sperm-egg fusion.

The beta hairpin is a simple protein structural motif involving two beta strands that look like a hairpin. The motif consists of two strands that are adjacent in primary structure, oriented in an antiparallel direction, and linked by a short loop of two to five amino acids. Beta hairpins can occur in isolation or as part of a series of hydrogen bonded strands that collectively comprise a beta sheet.

The EF hand is a helix–loop–helix structural domain or motif found in a large family of calcium-binding proteins.
The Cys-loop ligand-gated ion channel superfamily is composed of nicotinic acetylcholine, GABAA, GABAA-ρ, glycine, 5-HT3, and zinc-activated (ZAC) receptors. These receptors are composed of five protein subunits which form a pentameric arrangement around a central pore. There are usually 2 alpha subunits and 3 other beta, gamma, or delta subunits (some consist of 5 alpha subunits). The name of the family refers to a characteristic loop formed by 13 highly conserved amino acids between two cysteine (Cys) residues, which form a disulfide bond near the N-terminal extracellular domain.
CD2 is a cell adhesion molecule found on the surface of T cells and natural killer (NK) cells. It has also been called T-cell surface antigen T11/Leu-5, LFA-2, LFA-3 receptor, erythrocyte receptor and rosette receptor.
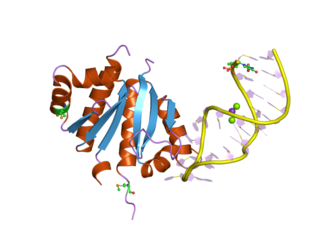
The K Homology (KH) domain is a protein domain that was first identified in the human heterogeneous nuclear ribonucleoprotein (hnRNP) K. An evolutionarily conserved sequence of around 70 amino acids, the KH domain is present in a wide variety of nucleic acid-binding proteins. The KH domain binds RNA, and can function in RNA recognition. It is found in multiple copies in several proteins, where they can function cooperatively or independently. For example, in the AU-rich element RNA-binding protein KSRP, which has 4 KH domains, KH domains 3 and 4 behave as independent binding modules to interact with different regions of the AU-rich RNA targets. The solution structure of the first KH domain of FMR1 and of the C-terminal KH domain of hnRNP K determined by nuclear magnetic resonance (NMR) revealed a beta-alpha-alpha-beta-beta-alpha structure. Autoantibodies to NOVA1, a KH domain protein, cause paraneoplastic opsoclonus ataxia. The KH domain is found at the N-terminus of the ribosomal protein S3. This domain is unusual in that it has a different fold compared to the normal KH domain.

FYN binding protein (FYB-120/130), also known as FYB, ADAP, and SLAP-130 is a protein that is encoded by the FYB gene in humans. The protein is expressed in T cells, monocytes, mast cells, macrophages, NK cells, but not B cells. FYB is a multifunctional protein involved in post-activation T cell signaling, lymphocyte cytokine production, cell adhesion, and actin remodeling.

Proline-serine-threonine phosphatase-interacting protein 1 is an enzyme that in humans is encoded by the PSTPIP1 gene.

CD2 antigen cytoplasmic tail-binding protein 2 is a protein that in humans is encoded by the CD2BP2 gene.
Coronin is an actin binding protein which also interacts with microtubules and in some cell types is associated with phagocytosis. Coronin proteins are expressed in a large number of eukaryotic organisms from yeast to humans.

WW domain-binding protein 1 is a protein that in humans is encoded by the WBP1 gene.
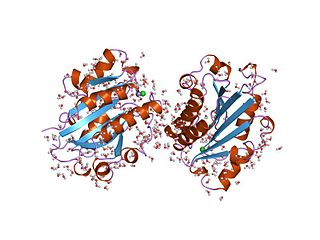
The von Willebrand factor type A (vWA) domain is a protein domain named after its occurrence in von Willebrand factor (vWF), a large multimeric glycoprotein found in blood plasma. Mutant forms of vWF are involved in the aetiology of bleeding disorders. This type A domain is the prototype for a protein superfamily.
The Walker A and Walker B motifs are protein sequence motifs, known to have highly conserved three-dimensional structures. These were first reported in ATP-binding proteins by Walker and co-workers in 1982.

The WW domain is a modular protein domain that mediates specific interactions with protein ligands. This domain is found in a number of unrelated signaling and structural proteins and may be repeated up to four times in some proteins. Apart from binding preferentially to proteins that are proline-rich, with particular proline-motifs, [AP]-P-P-[AP]-Y, some WW domains bind to phosphoserine- and phosphothreonine-containing motifs.
The thiol-activated Cholesterol-dependent Cytolysin(CDC) family is a member of the MACPF superfamily. Cholesterol dependent cytolysins are a family of β-barrel pore-forming exotoxins that are secreted by gram-positive bacteria. CDCs are secreted as water-soluble monomers of 50-70 kDa, that when bound to the target cell, form a circular homo-oligomeric complex containing as many as 40 monomers. Through multiple conformational changes, the β-barrel transmembrane structure is formed and inserted into the target cell membrane. The presence of cholesterol in the target membrane is required for pore formation, though the presence of cholesterol is not required by all CDCs for binding. For example, intermedilysin secreted by Streptococcus intermedius will bind only to target membranes containing a specific protein receptor, independent of the presence of cholesterol, but cholesterol is required by intermedilysin for pore formation. While the lipid environment of cholesterol in the membrane can affect toxin binding, the exact molecular mechanism that cholesterol regulates the cytolytic activity of the CDC is not fully understood.
The CHASE domain is an extracellular protein domain, which is found in transmembrane receptor from bacteria, lower eukaryotes and plants. It has been named CHASE because of its presence in diverse receptor-like proteins with histidine kinase and nucleotide cyclase domains. The CHASE domain is 200-230 amino acids long and always occurs N-terminally in extracellular or periplasmic locations, followed by an intracellular tail housing diverse enzymatic signalling domains such as histidine kinase, adenyl cyclase, GGDEF-type nucleotide cyclase and EAL-type phosphodiesterase domains, as well as non-enzymatic domains such PAS, GAF, phosphohistidine and response regulatory domains. The CHASE domain is predicted to bind diverse low molecular weight ligands, such as the cytokinin-like adenine derivatives or peptides, and mediate signal transduction through the respective receptors.
WH1 domain is an evolutionary conserved protein domain. Therefore, it has an important function.
UPF0575 protein C19orf67 is a protein which in humans is encoded by the C19orf67 gene. Orthologs of C19orf67 are found in many mammals, some reptiles, and most jawed fish. The protein is expressed at low levels throughout the body with the exception of the testis and breast tissue. Where it is expressed, the protein is predicted to be localized in the nucleus to carry out a function. The highly conserved and slowly evolving DUFF3314 region is predicted to form numerous alpha helices and may be vital to the function of the protein.













