
The mammalian target of rapamycin (mTOR), also referred to as the mechanistic target of rapamycin, and sometimes called FK506-binding protein 12-rapamycin-associated protein 1 (FRAP1), is a kinase that in humans is encoded by the MTOR gene. mTOR is a member of the phosphatidylinositol 3-kinase-related kinase family of protein kinases.
Eukaryotic translation is the biological process by which messenger RNA is translated into proteins in eukaryotes. It consists of four phases: initiation, elongation, termination, and recapping.
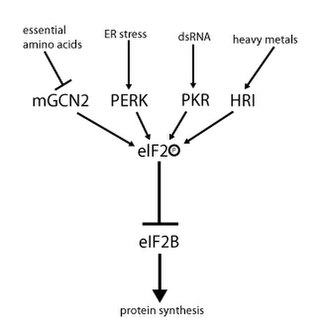
eIF-2-alpha kinase is a kinase enzyme that phosphorylates eIF2α. There are four forms in mammals:

Protein kinase RNA-activated also known as protein kinase R (PKR), interferon-induced, double-stranded RNA-activated protein kinase, or eukaryotic translation initiation factor 2-alpha kinase 2 (EIF2AK2) is an enzyme that in humans is encoded by the EIF2AK2 gene on chromosome 2. PKR is a serine/tyrosine kinase that is 551 amino acids long.
Eukaryotic initiation factors (eIFs) are proteins or protein complexes involved in the initiation phase of eukaryotic translation. These proteins help stabilize the formation of ribosomal preinitiation complexes around the start codon and are an important input for post-transcription gene regulation. Several initiation factors form a complex with the small 40S ribosomal subunit and Met-tRNAiMet called the 43S preinitiation complex. Additional factors of the eIF4F complex recruit the 43S PIC to the five-prime cap structure of the mRNA, from which the 43S particle scans 5'-->3' along the mRNA to reach an AUG start codon. Recognition of the start codon by the Met-tRNAiMet promotes gated phosphate and eIF1 release to form the 48S preinitiation complex, followed by large 60S ribosomal subunit recruitment to form the 80S ribosome. There exist many more eukaryotic initiation factors than prokaryotic initiation factors, reflecting the greater biological complexity of eukaryotic translation. There are at least twelve eukaryotic initiation factors, composed of many more polypeptides, and these are described below.
Eukaryotic translation initiation factor 4E-binding protein 1 is a protein that in humans is encoded by the EIF4EBP1 gene. inhibits cap-dependent translation by binding to translation initiation factor eIF4E. Phosphorylation of 4E-BP1 results in its release from eIF4E, thereby allows cap-dependent translation to continue thereby increasing the rate of protein synthesis.
RHEB also known as Ras homolog enriched in brain (RHEB) is a GTP-binding protein that is ubiquitously expressed in humans and other mammals. The protein is largely involved in the mTOR pathway and the regulation of the cell cycle.

DNA damage-inducible transcript 3, also known as C/EBP homologous protein (CHOP), is a pro-apoptotic transcription factor that is encoded by the DDIT3 gene. It is a member of the CCAAT/enhancer-binding protein (C/EBP) family of DNA-binding transcription factors. The protein functions as a dominant-negative inhibitor by forming heterodimers with other C/EBP members, preventing their DNA binding activity. The protein is implicated in adipogenesis and erythropoiesis and has an important role in the cell's stress response.

Eukaryotic translation initiation factor 2 subunit 1 (eIF2α) is a protein that in humans is encoded by the EIF2S1 gene.

Eukaryotic translation initiation factor 2-alpha kinase 3, also known as protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK), is an enzyme that in humans is encoded by the EIF2AK3 gene.

Eukaryotic translation initiation factor 2 subunit 2 (eIF2β) is a protein that in humans is encoded by the EIF2S2 gene.

Eukaryotic translation initiation factor 2A (eIF2A) is a protein that in humans is encoded by the EIF2A gene. The eIF2A protein is not to be confused with eIF2α, a subunit of the heterotrimeric eIF2 complex. Instead, eIF2A functions by a separate mechanism in eukaryotic translation.
52 kDa repressor of the inhibitor of the protein kinase is an enzyme that in humans is encoded by the PRKRIR gene.

Translational activator GCN1 is a protein that in humans is encoded by the GCN1L1 gene.
Eukaryotic Initiation Factor 2 (eIF2) is an eukaryotic initiation factor. It is required for most forms of eukaryotic translation initiation. eIF2 mediates the binding of tRNAiMet to the ribosome in a GTP-dependent manner. eIF2 is a heterotrimer consisting of an alpha, a beta, and a gamma subunit.
An upstream open reading frame (uORF) is an open reading frame (ORF) within the 5' untranslated region (5'UTR) of an mRNA. uORFs can regulate eukaryotic gene expression. Translation of the uORF typically inhibits downstream expression of the primary ORF. However, in some genes such as yeast GCN4, translation of specific uORFs may increase translation of the main ORF.
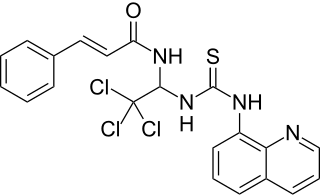
Salubrinal is a drug which acts as a specific inhibitor of eIF2α phosphatase enzymes and is primarily used experimentally, to study stress responses in eukaryotic cells associated with the action of eIF2. Salubrinal indirectly inhibits eIF2 as a result of reduced dephosphorylation of its α-subunit, resulting in activation of stress response pathways usually triggered by events such as oxidative stress or buildup of unfolded protein in the endoplasmic reticulum. Salubrinal has putative therapeutic value due to its function, but is as yet only used experimentally. Salubrinal is being studied at Indiana University for its potential to fight osteoporosis and accelerate bone healing.
Amino acid response is the mechanism triggered in mammalian cells by amino acid starvation.
The integrated stress response is a cellular stress response conserved in eukaryotic cells that downregulates protein synthesis and upregulates specific genes in response to internal or environmental stresses.
The transactivation domain or trans-activating domain (TAD) is a transcription factor scaffold domain which contains binding sites for other proteins such as transcription coregulators. These binding sites are frequently referred to as activation functions (AFs). TADs are named after their amino acid composition. These amino acids are either essential for the activity or simply the most abundant in the TAD. Transactivation by the Gal4 transcription factor is mediated by acidic amino acids, whereas hydrophobic residues in Gcn4 play a similar role. Hence, the TADs in Gal4 and Gcn4 are referred to as acidic or hydrophobic, respectively.













