
In molecular biology, a transcription factor (TF) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are expressed in the desired cells at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are 1500-1600 TFs in the human genome. Transcription factors are members of the proteome as well as regulome.

A homeobox is a DNA sequence, around 180 base pairs long, that regulates large-scale anatomical features in the early stages of embryonic development. Mutations in a homeobox may change large-scale anatomical features of the full-grown organism.

A zinc finger is a small protein structural motif that is characterized by the coordination of one or more zinc ions (Zn2+) in order to stabilize the fold. It was originally coined to describe the finger-like appearance of a hypothesized structure from the African clawed frog (Xenopus laevis) transcription factor IIIA. However, it has been found to encompass a wide variety of differing protein structures in eukaryotic cells. Xenopus laevis TFIIIA was originally demonstrated to contain zinc and require the metal for function in 1983, the first such reported zinc requirement for a gene regulatory protein followed soon thereafter by the Krüppel factor in Drosophila. It often appears as a metal-binding domain in multi-domain proteins.

DNA-binding proteins are proteins that have DNA-binding domains and thus have a specific or general affinity for single- or double-stranded DNA. Sequence-specific DNA-binding proteins generally interact with the major groove of B-DNA, because it exposes more functional groups that identify a base pair. However, there are some known minor groove DNA-binding ligands such as netropsin, distamycin, Hoechst 33258, pentamidine, DAPI and others.

Helix-turn-helix is a DNA-binding protein (DBP). The helix-turn-helix (HTH) is a major structural motif capable of binding DNA. Each monomer incorporates two α helices, joined by a short strand of amino acids, that bind to the major groove of DNA. The HTH motif occurs in many proteins that regulate gene expression. It should not be confused with the helix–loop–helix motif.
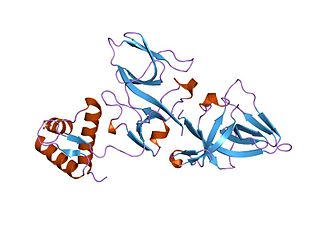
Repressor LexA or LexA is a transcriptional repressor that represses SOS response genes coding primarily for error-prone DNA polymerases, DNA repair enzymes and cell division inhibitors. LexA forms de facto a two-component regulatory system with RecA, which senses DNA damage at stalled replication forks, forming monofilaments and acquiring an active conformation capable of binding to LexA and causing LexA to cleave itself, in a process called autoproteolysis.

A basic helix–loop–helix (bHLH) is a protein structural motif that characterizes one of the largest families of dimerizing transcription factors. The word "basic" does not refer to complexity but to the chemistry of the motif because transcription factors in general contain basic amino acid residues in order to facilitate DNA binding.

A leucine zipper is a common three-dimensional structural motif in proteins. They were first described by Landschulz and collaborators in 1988 when they found that an enhancer binding protein had a very characteristic 30-amino acid segment and the display of these amino acid sequences on an idealized alpha helix revealed a periodic repetition of leucine residues at every seventh position over a distance covering eight helical turns. The polypeptide segments containing these periodic arrays of leucine residues were proposed to exist in an alpha-helical conformation and the leucine side chains from one alpha helix interdigitate with those from the alpha helix of a second polypeptide, facilitating dimerization.
A DNA-binding domain (DBD) is an independently folded protein domain that contains at least one structural motif that recognizes double- or single-stranded DNA. A DBD can recognize a specific DNA sequence or have a general affinity to DNA. Some DNA-binding domains may also include nucleic acids in their folded structure.

Tet Repressor proteins are proteins playing an important role in conferring antibiotic resistance to large categories of bacterial species.
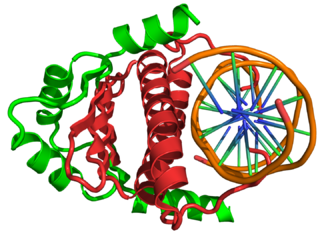
In the field of molecular biology, myocyte enhancer factor-2 (Mef2) proteins are a family of transcription factors which through control of gene expression are important regulators of cellular differentiation and consequently play a critical role in embryonic development. In adult organisms, Mef2 proteins mediate the stress response in some tissues. Mef2 proteins contain both MADS-box and Mef2 DNA-binding domains.

DNA-binding protein inhibitor ID-2 is a protein that in humans is encoded by the ID2 gene.
POU is a family of eukaryotic transcription factors that have well-conserved homeodomains. The Pou domain is a bipartite DNA binding domain found in these proteins.

ETS domain-containing protein Elk-4 is a protein that in humans is encoded by the ELK4 gene.
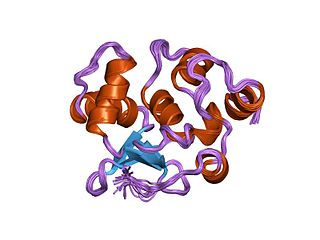
In the field of molecular biology, the ETSfamily is one of the largest families of transcription factors and is unique to animals. There are 29 genes in humans, 28 in the mouse, 10 in Caenorhabditis elegans and 9 in Drosophila. The founding member of this family was identified as a gene transduced by the leukemia virus, E26. The members of the family have been implicated in the development of different tissues as well as cancer progression.
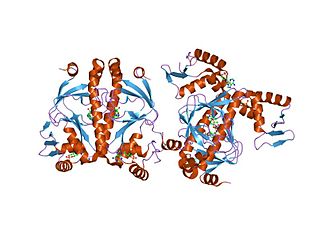
In molecular biology, the CRP domain is a protein domain consisting of a helix-turn-helix (HTH) motif. It is found at the C-terminus of numerous bacterial transcription regulatory proteins. These proteins bind DNA via the CRP domain. These proteins are very diverse, but for convenience may be grouped into subfamilies on the basis of sequence similarity. This family groups together a range of proteins, including ANR, CRP, CLP, CysR, FixK, Flp, FNR, FnrN, HlyX and NtcA.
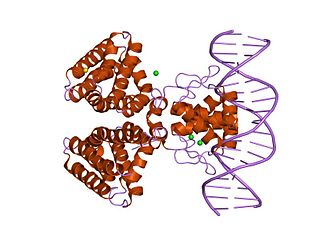
In molecular biology, the fatty acid metabolism regulator protein FadR, is a bacterial transcription factor.

In molecular biology, the iron dependent repressors are a family of bacterial and archaeal transcriptional repressors.

In molecular biology, the LuxR-type DNA-binding HTH domain is a DNA-binding, helix-turn-helix (HTH) domain of about 65 amino acids. It is present in transcription regulators of the LuxR/FixJ family of response regulators. The domain is named after Vibrio fischeri luxR, a transcriptional activator for quorum-sensing control of luminescence. LuxR-type HTH domain proteins occur in a variety of organisms. The DNA-binding HTH domain is usually located in the C-terminal region of the protein; the N-terminal region often containing an autoinducer-binding domain or a response regulatory domain. Most luxR-type regulators act as transcription activators, but some can be repressors or have a dual role for different sites. LuxR-type HTH regulators control a wide variety of activities in various biological processes.

A response regulator is a protein that mediates a cell's response to changes in its environment as part of a two-component regulatory system. Response regulators are coupled to specific histidine kinases which serve as sensors of environmental changes. Response regulators and histidine kinases are two of the most common gene families in bacteria, where two-component signaling systems are very common; they also appear much more rarely in the genomes of some archaea, yeasts, filamentous fungi, and plants. Two-component systems are not found in metazoans.


















