
The nucleoid is an irregularly shaped region within the prokaryotic cell that contains all or most of the genetic material. The chromosome of a typical prokaryote is circular, and its length is very large compared to the cell dimensions, so it needs to be compacted in order to fit. In contrast to the nucleus of a eukaryotic cell, it is not surrounded by a nuclear membrane. Instead, the nucleoid forms by condensation and functional arrangement with the help of chromosomal architectural proteins and RNA molecules as well as DNA supercoiling. The length of a genome widely varies and a cell may contain multiple copies of it.
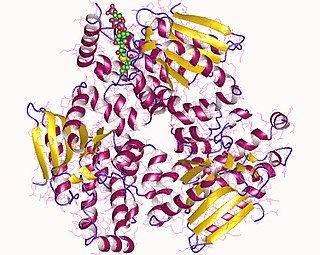
Enoyl-CoA-(∆) isomerase (EC 5.3.3.8, also known as dodecenoyl-CoA- isomerase, 3,2-trans-enoyl-CoA isomerase, ∆3 ,∆2 -enoyl-CoA isomerase, or acetylene-allene isomerase, is an enzyme that catalyzes the conversion of cis- or trans-double bonds of coenzyme A bound fatty acids at gamma-carbon to trans double bonds at beta-carbon as below:

Helix-turn-helix is a DNA-binding protein (DBP). The helix-turn-helix (HTH) is a major structural motif capable of binding DNA. Each monomer incorporates two α helices, joined by a short strand of amino acids, that bind to the major groove of DNA. The HTH motif occurs in many proteins that regulate gene expression. It should not be confused with the helix–loop–helix motif.

Acetyl-CoA carboxylase (ACC) is a biotin-dependent enzyme that catalyzes the irreversible carboxylation of acetyl-CoA to produce malonyl-CoA through its two catalytic activities, biotin carboxylase (BC) and carboxyltransferase (CT). ACC is a multi-subunit enzyme in most prokaryotes and in the chloroplasts of most plants and algae, whereas it is a large, multi-domain enzyme in the cytoplasm of most eukaryotes. The most important function of ACC is to provide the malonyl-CoA substrate for the biosynthesis of fatty acids. The activity of ACC can be controlled at the transcriptional level as well as by small molecule modulators and covalent modification. The human genome contains the genes for two different ACCs—ACACA and ACACB.
Propionyl-CoA is a coenzyme A derivative of propionic acid. It is composed of a 24 total carbon chain and its production and metabolic fate depend on which organism it is present in. Several different pathways can lead to its production, such as through the catabolism of specific amino acids or the oxidation of odd-chain fatty acids. It later can be broken down by propionyl-CoA carboxylase or through the methylcitrate cycle. In different organisms, however, propionyl-CoA can be sequestered into controlled regions, to alleviate its potential toxicity through accumulation. Genetic deficiencies regarding the production and breakdown of propionyl-CoA also have great clinical and human significance.
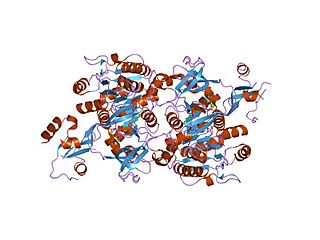
The long chain fatty acyl-CoA ligase is an enzyme of the ligase family that activates the oxidation of complex fatty acids. Long chain fatty acyl-CoA synthetase catalyzes the formation of fatty acyl-CoA by a two-step process proceeding through an adenylated intermediate. The enzyme catalyzes the following reaction,

Thiolases, also known as acetyl-coenzyme A acetyltransferases (ACAT), are enzymes which convert two units of acetyl-CoA to acetoacetyl CoA in the mevalonate pathway.
Pantothenate kinase (EC 2.7.1.33, PanK; CoaA) is the first enzyme in the Coenzyme A (CoA) biosynthetic pathway. It phosphorylates pantothenate (vitamin B5) to form 4'-phosphopantothenate at the expense of a molecule of adenosine triphosphate (ATP). It is the rate-limiting step in the biosynthesis of CoA.

DNA-binding protein inhibitor ID-2 is a protein that in humans is encoded by the ID2 gene.
In enzymology, a lipoyl(octanoyl) transferase (EC 2.3.1.181) is an enzyme that catalyzes the chemical reaction

C-terminal-binding protein 1 also known as CtBP1 is a protein that in humans is encoded by the CTBP1 gene. CtBP1 is one of two CtBP proteins, the other protein being CtBP2.

DNA-binding protein inhibitor ID-3 is a protein that in humans is encoded by the ID3 gene.

Nuclear inhibitor of protein phosphatase 1 is an enzyme that in humans is encoded by the PPP1R8 gene.

Fatty acid desaturase 1 (FADS1) is an enzyme that in humans is encoded by the FADS1 gene.

The human ETFB gene encodes the Electron-transfer-flavoprotein, beta subunit, also known as ETF-β. Together with Electron-transfer-flavoprotein, alpha subunit, encoded by the 'ETFA' gene, it forms the heterodimeric Electron transfer flavoprotein (ETF). The native ETF protein contains one molecule of FAD and one molecule of AMP, respectively.
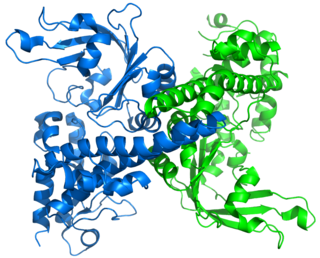
Restriction endonuclease (REase) EcoRII is an enzyme of restriction modification system (RM) naturally found in Escherichia coli, a Gram-negative bacteria. Its molecular mass is 45.2 kDa, being composed of 402 amino acids.

In molecular biology, the ars operon is an operon found in several bacterial taxon. It is required for the detoxification of arsenate, arsenite, and antimonite. This system transports arsenite and antimonite out of the cell. The pump is composed of two polypeptides, the products of the arsA and arsB genes. This two-subunit enzyme produces resistance to arsenite and antimonite. Arsenate, however, must first be reduced to arsenite before it is extruded. A third gene, arsC, expands the substrate specificity to allow for arsenate pumping and resistance. ArsC is an approximately 150-residue arsenate reductase that uses reduced glutathione (GSH) to convert arsenate to arsenite with a redox active cysteine residue in the active site. ArsC forms an active quaternary complex with GSH, arsenate, and glutaredoxin 1 (Grx1). The three ligands must be present simultaneously for reduction to occur.

In molecular biology, the LuxR-type DNA-binding HTH domain is a DNA-binding, helix-turn-helix (HTH) domain of about 65 amino acids. It is present in transcription regulators of the LuxR/FixJ family of response regulators. The domain is named after Vibrio fischeri luxR, a transcriptional activator for quorum-sensing control of luminescence. LuxR-type HTH domain proteins occur in a variety of organisms. The DNA-binding HTH domain is usually located in the C-terminal region of the protein; the N-terminal region often containing an autoinducer-binding domain or a response regulatory domain. Most luxR-type regulators act as transcription activators, but some can be repressors or have a dual role for different sites. LuxR-type HTH regulators control a wide variety of activities in various biological processes.
EnvZ/OmpR is a two-component regulatory system widely distributed in bacteria and particularly well characterized in Escherichia coli. Its function is in osmoregulation, responding to changes in environmental osmolality by regulating the expression of the outer membrane porins OmpF and OmpC. EnvZ is a histidine kinase which also possesses a cytoplasmic osmosensory domain, and OmpR is its corresponding response regulator protein.

In molecular biology, the GntR-like bacterial transcription factors are a family of transcription factors.
















