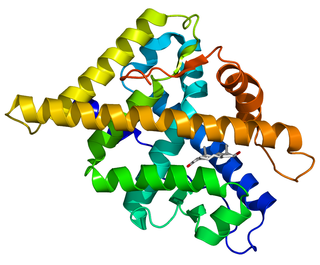
The androgen receptor (AR), also known as NR3C4, is a type of nuclear receptor that is activated by binding any of the androgenic hormones, including testosterone and dihydrotestosterone, in the cytoplasm and then translocating into the nucleus. The androgen receptor is most closely related to the progesterone receptor, and progestins in higher dosages can block the androgen receptor.

The death-effector domain (DED) is a protein interaction domain found only in eukaryotes that regulates a variety of cellular signalling pathways. The DED domain is found in inactive procaspases and proteins that regulate caspase activation in the apoptosis cascade such as FAS-associating death domain-containing protein (FADD). FADD recruits procaspase 8 and procaspase 10 into a death induced signaling complex (DISC). This recruitment is mediated by a homotypic interaction between the procaspase DED and a second DED that is death effector domain in an adaptor protein that is directly associated with activated TNF receptors. Complex formation allows proteolytic activation of procaspase into the active caspase form which results in the initiation of apoptosis. Structurally the DED domain are a subclass of protein motif known as the death fold and contains 6 alpha helices, that closely resemble the structure of the Death domain (DD).

In the field of cell biology, TNF-related apoptosis-inducing ligand (TRAIL), is a protein functioning as a ligand that induces the process of cell death called apoptosis.

Huntingtin(Htt) is the protein coded for in humans by the HTT gene, also known as the IT15 ("interesting transcript 15") gene. Mutated HTT is the cause of Huntington's disease (HD), and has been investigated for this role and also for its involvement in long-term memory storage.

The p75 neurotrophin receptor (p75NTR) was first identified in 1973 as the low-affinity nerve growth factor receptor (LNGFR) before discovery that p75NTR bound other neurotrophins equally well as nerve growth factor. p75NTR is a neurotrophic factor receptor. Neurotrophic factor receptors bind Neurotrophins including Nerve growth factor, Neurotrophin-3, Brain-derived neurotrophic factor, and Neurotrophin-4. All neurotrophins bind to p75NTR. This also includes the immature pro-neurotrophin forms. Neurotrophic factor receptors, including p75NTR, are responsible for ensuring a proper density to target ratio of developing neurons, refining broader maps in development into precise connections. p75NTR is involved in pathways that promote neuronal survival and neuronal death.

The luteinizing hormone/choriogonadotropin receptor (LHCGR), also lutropin/choriogonadotropin receptor (LCGR) or luteinizing hormone receptor (LHR) is a transmembrane receptor found predominantly in the ovary and testis, but also many extragonadal organs such as the uterus and breasts. The receptor interacts with both luteinizing hormone (LH) and chorionic gonadotropins and represents a G protein-coupled receptor (GPCR). Its activation is necessary for the hormonal functioning during reproduction.

Cyclic adenosine monophosphate Response Element Binding protein Binding Protein, also known as CREBBP or CBP or KAT3A, is a coactivator encoded by the CREBBP gene in humans, located on chromosome 16p13.3. CBP has intrinsic acetyltransferase functions; it is able to add acetyl groups to both transcription factors as well as histone lysines, the latter of which has been shown to alter chromatin structure making genes more accessible for transcription. This relatively unique acetyltransferase activity is also seen in another transcription enzyme, EP300 (p300). Together, they are known as the p300-CBP coactivator family and are known to associate with more than 16,000 genes in humans; however, while these proteins share many structural features, emerging evidence suggests that these two co-activators may promote transcription of genes with different biological functions.

Caspase-8 is a caspase protein, encoded by the CASP8 gene. It most likely acts upon caspase-3. CASP8 orthologs have been identified in numerous mammals for which complete genome data are available. These unique orthologs are also present in birds.

X-linked inhibitor of apoptosis protein (XIAP), also known as inhibitor of apoptosis protein 3 (IAP3) and baculoviral IAP repeat-containing protein 4 (BIRC4), is a protein that stops apoptotic cell death. In humans, this protein (XIAP) is produced by a gene named XIAP gene located on the X chromosome.
Huntingtin-associated protein 1 (HAP1) is a protein which in humans is encoded by the HAP1 gene. This protein was found to bind to the mutant huntingtin protein (mHtt) in proportion to the number of glutamines present in the glutamine repeat region.

ERG is an oncogene. ERG is a member of the ETS family of transcription factors. The ERG gene encodes for a protein, also called ERG, that functions as a transcriptional regulator. Genes in the ETS family regulate embryonic development, cell proliferation, differentiation, angiogenesis, inflammation, and apoptosis.

Hematopoietic lineage cell-specific protein is a protein that in humans is encoded by the HCLS1 gene.

Cdc42-interacting protein 4 is a protein that in humans is encoded by the TRIP10 gene.

Deleted in Liver Cancer 1 also known as DLC1 and StAR-related lipid transfer protein 12 (STARD12) is a protein which in humans is encoded by the DLC1 gene.

Histone acetyltransferase KAT7 is an enzyme that in humans is encoded by the KAT7 gene. It specifically acetylates H4 histones at the lysine12 residue (H4K12) and is necessary for origin licensing and DNA replication. KAT7 associates with origins of replication during G1 phase of the cell cycle through complexing with CDT1. Geminin is thought to inhibit the acetyltransferase activity of KAT7 when KAT7 and CDT1 are complexed together.

Transcription factor IIIA is a protein that in humans is encoded by the GTF3A gene. It was first isolated and characterized by Wolffe and Brown in 1988.

CASP8-associated protein 2 is a protein, that in humans is encoded by the CASP8AP2 gene.
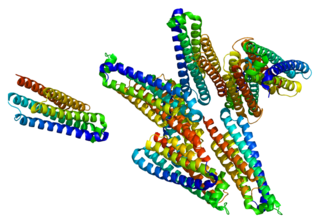
Huntingtin-interacting protein 1-related protein is a protein that in humans is encoded by the HIP1R gene.

Intraflagellar transport protein 57 homolog is a protein that in humans is encoded by the IFT57 gene.

Palmitoyltransferase ZDHHC17 is an enzyme that contains a DHHC domain that in humans is encoded by the ZDHHC17 gene.






















