Related Research Articles

In immunology, an antigen (Ag) is a molecule, moiety, foreign particulate matter, or an allergen, such as pollen, that can bind to a specific antibody or T-cell receptor. The presence of antigens in the body may trigger an immune response.

Natural killer cells, also known as NK cells or large granular lymphocytes (LGL), are a type of cytotoxic lymphocyte critical to the innate immune system that belong to the rapidly expanding family of known innate lymphoid cells (ILC) and represent 5–20% of all circulating lymphocytes in humans. The role of NK cells is analogous to that of cytotoxic T cells in the vertebrate adaptive immune response. NK cells provide rapid responses to virus-infected cell and other intracellular pathogens acting at around 3 days after infection, and respond to tumor formation. Typically, immune cells detect the antigen presented on major histocompatibility complex (MHC) on infected cell surfaces, triggering cytokine release, causing the death of the infected cell by lysis or apoptosis. NK cells are unique, however, as they have the ability to recognize and kill stressed cells in the absence of antibodies and MHC, allowing for a much faster immune reaction. They were named "natural killers" because of the notion that they do not require activation to kill cells that are missing "self" markers of MHC class I. This role is especially important because harmful cells that are missing MHC I markers cannot be detected and destroyed by other immune cells, such as T lymphocyte cells.

The adaptive immune system, also known as the acquired immune system, or specific immune system is a subsystem of the immune system that is composed of specialized, systemic cells and processes that eliminate pathogens or prevent their growth. The acquired immune system is one of the two main immunity strategies found in vertebrates.
Antigen processing, or the cytosolic pathway, is an immunological process that prepares antigens for presentation to special cells of the immune system called T lymphocytes. It is considered to be a stage of antigen presentation pathways. This process involves two distinct pathways for processing of antigens from an organism's own (self) proteins or intracellular pathogens, or from phagocytosed pathogens ; subsequent presentation of these antigens on class I or class II major histocompatibility complex (MHC) molecules is dependent on which pathway is used. Both MHC class I and II are required to bind antigens before they are stably expressed on a cell surface. MHC I antigen presentation typically involves the endogenous pathway of antigen processing, and MHC II antigen presentation involves the exogenous pathway of antigen processing. Cross-presentation involves parts of the exogenous and the endogenous pathways but ultimately involves the latter portion of the endogenous pathway.

An antigen-presenting cell (APC) or accessory cell is a cell that displays antigen bound by major histocompatibility complex (MHC) proteins on its surface; this process is known as antigen presentation. T cells may recognize these complexes using their T cell receptors (TCRs). APCs process antigens and present them to T-cells.

MHC class I molecules are one of two primary classes of major histocompatibility complex (MHC) molecules and are found on the cell surface of all nucleated cells in the bodies of vertebrates. They also occur on platelets, but not on red blood cells. Their function is to display peptide fragments of proteins from within the cell to cytotoxic T cells; this will trigger an immediate response from the immune system against a particular non-self antigen displayed with the help of an MHC class I protein. Because MHC class I molecules present peptides derived from cytosolic proteins, the pathway of MHC class I presentation is often called cytosolic or endogenous pathway.
Cross-presentation is the ability of certain professional antigen-presenting cells (mostly dendritic cells) to take up, process and present extracellular antigens with MHC class I molecules to CD8 T cells (cytotoxic T cells). Cross-priming, the result of this process, describes the stimulation of naive cytotoxic CD8+ T cells into activated cytotoxic CD8+ T cells. This process is necessary for immunity against most tumors and against viruses that infect dendritic cells and sabotage their presentation of virus antigens. Cross presentation is also required for the induction of cytotoxic immunity by vaccination with protein antigens, for example, tumour vaccination.

Antigen presentation is a vital immune process that is essential for T cell immune response triggering. Because T cells recognize only fragmented antigens displayed on cell surfaces, antigen processing must occur before the antigen fragment, now bound to the major histocompatibility complex (MHC), is transported to the surface of the cell, a process known as presentation, where it can be recognized by a T-cell receptor. If there has been an infection with viruses or bacteria, the cell will present an endogenous or exogenous peptide fragment derived from the antigen by MHC molecules. There are two types of MHC molecules which differ in the behaviour of the antigens: MHC class I molecules (MHC-I) bind peptides from the cell cytosol, while peptides generated in the endocytic vesicles after internalisation are bound to MHC class II (MHC-II). Cellular membranes separate these two cellular environments - intracellular and extracellular. Each T cell can only recognize tens to hundreds of copies of a unique sequence of a single peptide among thousands of other peptides presented on the same cell, because an MHC molecule in one cell can bind to quite a large range of peptides. Predicting which antigens will be presented to the immune system by a certain MHC/HLA type is difficult, but the technology involved is improving.

MHC Class II molecules are a class of major histocompatibility complex (MHC) molecules normally found only on professional antigen-presenting cells such as dendritic cells, mononuclear phagocytes, some endothelial cells, thymic epithelial cells, and B cells. These cells are important in initiating immune responses.
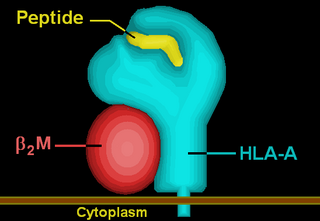
HLA-A is a group of human leukocyte antigens (HLA) that are encoded by the HLA-A locus, which is located at human chromosome 6p21.3. HLA is a major histocompatibility complex (MHC) antigen specific to humans. HLA-A is one of three major types of human MHC class I transmembrane proteins. The others are HLA-B and HLA-C. The protein is a heterodimer, and is composed of a heavy α chain and smaller β chain. The α chain is encoded by a variant HLA-A gene, and the β chain (β2-microglobulin) is an invariant β2 microglobulin molecule. The β2 microglobulin protein is encoded by the B2M gene, which is located at chromosome 15q21.1 in humans.

Minor histocompatibility antigen are peptides presented on the cellular surface of donated organs that are known to give an immunological response in some organ transplants. They cause problems of rejection less frequently than those of the major histocompatibility complex (MHC). Minor histocompatibility antigens (MiHAs) are diverse, short segments of proteins and are referred to as peptides. These peptides are normally around 9-12 amino acids in length and are bound to both the major histocompatibility complex (MHC) class I and class II proteins. Peptide sequences can differ among individuals and these differences arise from SNPs in the coding region of genes, gene deletions, frameshift mutations, or insertions. About a third of the characterized MiHAs come from the Y chromosome. Prior to becoming a short peptide sequence, the proteins expressed by these polymorphic or diverse genes need to be digested in the proteasome into shorter peptides. These endogenous or self peptides are then transported into the endoplasmic reticulum with a peptide transporter pump called TAP where they encounter and bind to the MHC class I molecule. This contrasts with MHC class II molecules's antigens which are peptides derived from phagocytosis/endocytosis and molecular degradation of non-self entities' proteins, usually by antigen-presenting cells. MiHA antigens are either ubiquitously expressed in most tissue like skin and intestines or restrictively expressed in the immune cells.
Ly49 is a family of membrane C-type lectin-like receptors expressed mainly on NK cells but also on other immune cells. Their primary role is to bind MHC-I molecules to distinguish between self healthy cells and infected or altered cells. Ly49 family is coded by Klra gene cluster and include genes for both inhibitory and activating paired receptors, but most of them are inhibitory. Inhibitory Ly49 receptors play a role in the recognition of self cells and thus maintain self-tolerance and prevent autoimmunity by suppressing NK cell activation. On the other hand, activating receptors recognise ligands from cancer or viral infected cells and are used when cells lack or have abnormal expression of MHC-I molecules, which activate cytokine production and cytotoxic activity of NK and immune cells.
NKG2 also known as CD159 is a receptor for natural killer cells. There are 7 NKG2 types: A, B, C, D, E, F and H. NKG2D is an activating receptor on the NK cell surface. NKG2A dimerizes with CD94 to make an inhibitory receptor (CD94/NKG2).

HLA-DM is an intracellular protein involved in the mechanism of antigen presentation on antigen presenting cells (APCs) of the immune system. It does this by assisting in peptide loading of major histocompatibility complex (MHC) class II membrane-bound proteins. HLA-DM is encoded by the genes HLA-DMA and HLA-DMB.

UL16 binding protein 2 (ULBP2) is a cell surface glycoprotein encoded by ULBP2 gene located on the chromosome 6. ULBP2 is related to MHC class I molecules, but its gene maps outside the MHC locus. The domain structure of ULBP2 differs significantly from those of conventional MHC class I molecules. It does not contain the α3 domain and the transmembrane segment. ULBP2 is thus composed of only the α1α2 domain which is linked to the cell membrane by the GPI anchor.

UL16 binding protein 1 (ULBP1) is a cell surface glycoprotein encoded by ULBP1 gene located on the chromosome 6. ULBP1 is related to MHC class I molecules, but its gene maps outside the MHC locus. The domain structure of ULBP1 differs significantly from those of conventional MHC class I molecules. It does not contain the α3 domain and the transmembrane segment. ULBP1 is thus composed of only the α1α2 domain which is linked to the cell membrane by the GPI anchor. It functions as a stress-induced ligand for NKG2D receptor. ULBP1 is, for example, upregulated during HCMV infection. Binding of HCMV-encoded UL16 glycoprotein to ULBP1 interferes with cell surface localization of ULBP1; this represents another mechanism by which HCMV-infected cells might escape the immune system.

NKG2D is an activating receptor (transmembrane protein) belonging to the NKG2 family of C-type lectin-like receptors. NKG2D is encoded by KLRK1 (killer cell lectin like receptor K1) gene which is located in the NK-gene complex (NKC) situated on chromosome 6 in mice and chromosome 12 in humans. In mice, it is expressed by NK cells, NK1.1+ T cells, γδ T cells, activated CD8+ αβ T cells and activated macrophages. In humans, it is expressed by NK cells, γδ T cells and CD8+ αβ T cells. NKG2D recognizes induced-self proteins from MIC and RAET1/ULBP families which appear on the surface of stressed, malignant transformed, and infected cells.
Induced-self antigen is a marker of abnormal self, which can be recognized upon infected and transformed cells. Therefore, the recognition of "induced self" is an important strategy for surveillance of infection or tumor transformation - it results in elimination of the affected cells by activated NK cells or other immunological mechanisms. Similarly γδ T cells can recognize induced-self antigens expressed on cells under stress conditions.
CD94/NKG2 is a family of C-type lectin receptors which are expressed predominantly on the surface of NK cells and a subset of CD8+ T-lymphocyte. These receptors stimulate or inhibit cytotoxic activity of NK cells, therefore they are divided into activating and inhibitory receptors according to their function. CD94/NKG2 recognize nonclassical MHC glycoproteins class I (HLA-E in human and Qa-1 molecules in the mouse).

The peptide-loading complex (PLC) is a short-lived, multisubunit membrane protein complex that is located in the endoplasmic reticulum (ER). It orchestrates peptide translocation and selection by major histocompatibility complex class I (MHC-I) molecules. Stable peptide-MHC I complexes are released to the cell surface to promote T-cell response against malignant or infected cells. In turn, T-cells recognize the activated peptides, which could be immunogenic or non-immunogenic.
References
- 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 Lilley, Brendan N.; Ploegh, Hidde L. (2005). "Viral modulation of antigen presentation: manipulation of cellular targets in the ER and beyond". Immunological Reviews. 207 (1): 126–144. doi:10.1111/j.0105-2896.2005.00318.x. ISSN 1600-065X. PMID 16181332. S2CID 14977488.
- 1 2 Berry, Richard; Watson, Gabrielle M.; Jonjic, Stipan; Degli-Esposti, Mariapia A.; Rossjohn, Jamie (February 2020). "Modulation of innate and adaptive immunity by cytomegaloviruses". Nature Reviews Immunology. 20 (2): 113–127. doi:10.1038/s41577-019-0225-5. ISSN 1474-1733. PMID 31666730. S2CID 204942568.
- 1 2 3 4 Zeleznjak, Jelena; Popovic, Branka; Krmpotic, Astrid; Jonjic, Stipan; Lisnic, Vanda Juranic (2017-09-01). "Mouse cytomegalovirus encoded immunoevasins and evolution of Ly49 receptors – Sidekicks or enemies?". Immunology Letters. 3rd meeting of Middle-European Societies for Immunology and Allergology. 189: 40–47. doi:10.1016/j.imlet.2017.04.007. ISSN 0165-2478. PMID 28414184.
- ↑ Janeway. Immuno Biology. Garland Science, Taylor & Francis Group, LLC, 2008. p. 189–190.
- 1 2 3 4 5 6 7 8 9 10 11 12 van de Weijer, Michael L.; Luteijn, Rutger D.; Wiertz, Emmanuel J. H. J. (2015-03-01). "Viral immune evasion: Lessons in MHC class I antigen presentation". Seminars in Immunology. What do pathogens teach us about the immune system?. 27 (2): 125–137. doi:10.1016/j.smim.2015.03.010. ISSN 1044-5323. PMID 25887630.
- 1 2 3 4 5 Loureiro, Joana; Ploegh, Hidde L. (2006). Antigen Presentation and the Ubiquitin‐Proteasome System in Host–Pathogen Interactions. Advances in Immunology. Vol. 92. pp. 225–305. doi:10.1016/S0065-2776(06)92006-9. ISBN 9780123736369. ISSN 0065-2776. PMC 7112114 . PMID 17145306.
- 1 2 3 Gewurz, Benjamin E; Gaudet, Rachelle; Tortorella, Domenico; Wang, Evelyn W; Ploegh, Hidde L (2001-08-01). "Virus subversion of immunity: a structural perspective". Current Opinion in Immunology. 13 (4): 442–450. doi:10.1016/S0952-7915(00)00239-9. ISSN 0952-7915. PMID 11498300.
- 1 2 3 4 Reddehase, Matthias J. (November 2002). "Antigens and immunoevasins: opponents in cytomegalovirus immune surveillance". Nature Reviews Immunology. 2 (11): 831–844. doi: 10.1038/nri932 . ISSN 1474-1741. PMID 12415307. S2CID 8698447.
- 1 2 Li, Yili; Mariuzza, Roy A. (2014-03-26). "Structural Basis for Recognition of Cellular and Viral Ligands by NK Cell Receptors". Frontiers in Immunology. 5: 123. doi: 10.3389/fimmu.2014.00123 . ISSN 1664-3224. PMC 3972465 . PMID 24723923.
- 1 2 Reddehase, Matthias J; Simon, Christian O; Podlech, Jürgen; Holtappels, Rafaela (2004-05-01). "Stalemating a clever opportunist: lessons from murine cytomegalovirus". Human Immunology. 65 (5): 446–455. doi:10.1016/j.humimm.2004.02.024. ISSN 0198-8859. PMID 15172444.