
Salmonella is a genus of rod-shaped (bacillus) gram-negative bacteria of the family Enterobacteriaceae. The two known species of Salmonella are Salmonella enterica and Salmonella bongori. S. enterica is the type species and is further divided into six subspecies that include over 2,650 serotypes. Salmonella was named after Daniel Elmer Salmon (1850–1914), an American veterinary surgeon.
Charles Yanofsky was an American geneticist on the faculty of Stanford University who contributed to the establishment of the one gene-one enzyme hypothesis and discovered attenuation, a riboswitch mechanism in which messenger RNA changes shape in response to a small molecule and thus alters its binding ability for the regulatory region of a gene or operon.

The trp operon is a group of genes that are transcribed together, encoding the enzymes that produce the amino acid tryptophan in bacteria. The trp operon was first characterized in Escherichia coli, and it has since been discovered in many other bacteria. The operon is regulated so that, when tryptophan is present in the environment, the genes for tryptophan synthesis are repressed.
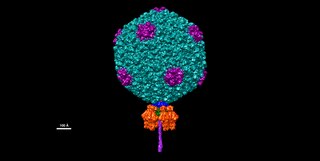
Salmonella virus P22 is a bacteriophage in the Podoviridae family that infects Salmonella typhimurium. Like many phages, it has been used in molecular biology to induce mutations in cultured bacteria and to introduce foreign genetic material. P22 has been used in generalized transduction and is an important tool for investigating Salmonella genetics.

Walter Dobrogosz is a professor emeritus of North Carolina State University, best known for his discovery and further research on the probiotic bacterium Lactobacillus reuteri.
The L-arabinose operon, also called the ara or araBAD operon, is an operon required for the breakdown of the five-carbon sugar L-arabinose in Escherichia coli. The L-arabinose operon contains three structural genes: araB, araA, araD, which encode for three metabolic enzymes that are required for the metabolism of L-arabinose. AraB (ribulokinase), AraA, and AraD produced by these genes catalyse conversion of L-arabinose to an intermediate of the pentose phosphate pathway, D-xylulose-5-phosphate.
In molecular genetics, a regulon is a group of genes that are regulated as a unit, generally controlled by the same regulatory gene that expresses a protein acting as a repressor or activator. This terminology is generally, although not exclusively, used in reference to prokaryotes, whose genomes are often organized into operons; the genes contained within a regulon are usually organized into more than one operon at disparate locations on the chromosome. Applied to eukaryotes, the term refers to any group of non-contiguous genes controlled by the same regulatory gene.
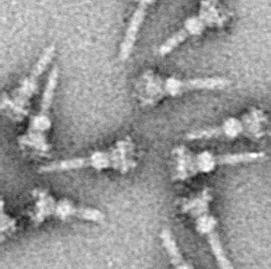
The type III secretion system is one of the bacterial secretion systems used by bacteria to secrete their effector proteins into the host's cells to promote virulence and colonisation. While the type III secretion system has been widely regarded as equivalent to the injectisome, many argue that the injectisome is only part of the type III secretion system, which also include structures like the flagellar export apparatus. The T3SS is a needle-like protein complex found in several species of pathogenic gram-negative bacteria.

The gcvB RNA gene encodes a small non-coding RNA involved in the regulation of a number of amino acid transport systems as well as amino acid biosynthetic genes. The GcvB gene is found in enteric bacteria such as Escherichia coli. GcvB regulates genes by acting as an antisense binding partner of the mRNAs for each regulated gene. This binding is dependent on binding to a protein called Hfq. Transcription of the GcvB RNA is activated by the adjacent GcvA gene and repressed by the GcvR gene. A deletion of GcvB RNA from Y. pestis changed colony shape as well as reducing growth. It has been shown by gene deletion that GcvB is a regulator of acid resistance in E. coli. GcvB enhances the ability of the bacterium to survive low pH by upregulating the levels of the alternate sigma factor RpoS. A polymeric form of GcvB has recently been identified. Interaction of GcvB with small RNA SroC triggers the degradation of GcvB by RNase E, lifting the GcvB-mediated mRNA repression of its target genes.

In enzymology, a precorrin-8X methylmutase is an enzyme that catalyzes the chemical reaction

In enzymology, a nicotinate-nucleotide-dimethylbenzimidazole phosphoribosyltransferase is an enzyme that catalyzes the chemical reaction

Leucine responsive protein, or Lrp, is a global regulator protein, meaning that it regulates the biosynthesis of leucine, as well as the other branched-chain amino acids, valine and isoleucine. In bacteria, it is encoded by the lrp gene.

Bacterial microcompartments (BMCs) are organelle-like structures found in bacteria. They consist of a protein shell that encloses enzymes and other proteins. BMCs are typically about 40–200 nanometers in diameter and are made entirely of proteins. The shell functions like a membrane, as it is selectively permeable. Other protein-based compartments found in bacteria and archaea include encapsulin nanocompartments and gas vesicles.

Naomi Datta, FRS was a distinguished British geneticist. Working at Hammersmith Hospital in the 1950s and early 1960s, she identified horizontal gene transfer as a source of multi-antibiotic resistance in bacteria.
Transposons are semi-parasitic DNA sequences which can replicate and spread through the host's genome. They can be harnessed as a genetic tool for analysis of gene and protein function. The use of transposons is well-developed in Drosophila and in Thale cress and bacteria such as Escherichia coli.

In molecular biology, cob(I)yrinic acid a,c-diamide adenosyltransferase EC 2.5.1.17 is an enzyme which catalyses the conversion of cobalamin into one of its coenzyme forms, adenosylcobalamin. Adenosylcobalamin is required as a cofactor for the activity of certain enzymes. AdoCbl contains an adenosyl moiety liganded to the cobalt ion of cobalamin via a covalent Co-C bond.
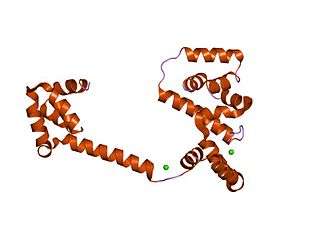
In molecular biology, the flagellar motor switch protein(Flig) is one of three proteins in certain bacteria coded for by the gene fliG. The other two proteins are FliN coded for by fliN, and FliM coded for by fliM. The protein complex regulates the direction of flagellar rotation and hence controls swimming behaviour. The switch is a complex apparatus that responds to signals transduced by the chemotaxis sensory signalling system during chemotactic behaviour. CheY, the chemotaxis response regulator, is believed to act directly on the switch to induce a switch in the flagellar motor direction of rotation.
The C4-dicarboxylate uptake family or Dcu family is a family of transmembrane ion transporters found in bacteria. Their function is to exchange dicarboxylates such as aspartate, malate, fumarate and succinate.
Transcription-translation coupling is a mechanism of gene expression regulation in which synthesis of an mRNA (transcription) is affected by its concurrent decoding (translation). In prokaryotes, mRNAs are translated while they are transcribed. This allows communication between RNA polymerase, the multisubunit enzyme that catalyzes transcription, and the ribosome, which catalyzes translation. Coupling involves both direct physical interactions between RNA polymerase and the ribosome, as well as ribosome-induced changes to the structure and accessibility of the intervening mRNA that affect transcription.
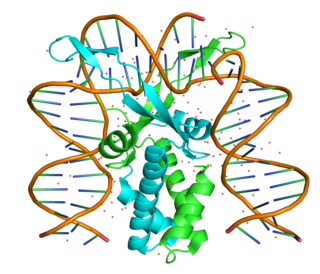
The integration host factor (IHF) is a bacterial DNA binding protein complex that facilitates genetic recombination, replication, and transcription by binding to specific DNA sequences and bending the DNA. It also facilitates the integration of foreign DNA into the host genome. It is a heterodimeric complex composed of two homologous subunits IHFalpha and IHFbeta.













