
Caenorhabditis elegans is a free-living transparent nematode about 1 mm in length that lives in temperate soil environments. It is the type species of its genus. The name is a blend of the Greek caeno- (recent), rhabditis (rod-like) and Latin elegans (elegant). In 1900, Maupas initially named it Rhabditides elegans. Osche placed it in the subgenus Caenorhabditis in 1952, and in 1955, Dougherty raised Caenorhabditis to the status of genus.
A genetic screen or mutagenesis screen is an experimental technique used to identify and select individuals who possess a phenotype of interest in a mutagenized population. Hence a genetic screen is a type of phenotypic screen. Genetic screens can provide important information on gene function as well as the molecular events that underlie a biological process or pathway. While genome projects have identified an extensive inventory of genes in many different organisms, genetic screens can provide valuable insight as to how those genes function.
Gene knockdown is an experimental technique by which the expression of one or more of an organism's genes is reduced. The reduction can occur either through genetic modification or by treatment with a reagent such as a short DNA or RNA oligonucleotide that has a sequence complementary to either gene or an mRNA transcript.

Sir John Edward Sulston was a British biologist and academic who won the Nobel Prize in Physiology or Medicine for his work on the cell lineage and genome of the worm Caenorhabditis elegans in 2002 with his colleagues Sydney Brenner and Robert Horvitz at the MRC Laboratory of Molecular Biology. He was a leader in human genome research and Chair of the Institute for Science, Ethics and Innovation at the University of Manchester. Sulston was in favour of science in the public interest, such as free public access of scientific information and against the patenting of genes and the privatisation of genetic technologies.
The DAF-2 gene encodes for the insulin-like growth factor 1 (IGF-1) receptor in the worm Caenorhabditis elegans. DAF-2 is part of the first metabolic pathway discovered to regulate the rate of aging. DAF-2 is also known to regulate reproductive development, resistance to oxidative stress, thermotolerance, resistance to hypoxia, and resistance to bacterial pathogens. Mutations in DAF-2 and also Age-1 have been shown by Cynthia Kenyon to double the lifespan of the worms. In a 2007 episode of WNYC’s Radiolab, Kenyon called DAF-2 "the grim reaper gene.”
John Graham White is an Emeritus Professor of Anatomy and Molecular Biology at the University of Wisconsin–Madison. His research interests are in the biology of the model organism Caenorhabditis elegans and laser microscopy.

Most animal testing involves invertebrates, especially Drosophila melanogaster, a fruit fly, and Caenorhabditis elegans, a nematode. These animals offer scientists many advantages over vertebrates, including their short life cycle, simple anatomy and the ease with which large numbers of individuals may be studied. Invertebrates are often cost-effective, as thousands of flies or nematodes can be housed in a single room.

Cornelia Isabella "Cori" Bargmann is an American neurobiologist. She is known for her work on the genetic and neural circuit mechanisms of behavior using C. elegans, particularly the mechanisms of olfaction in the worm. She has been elected to the National Academy of Sciences and had been a Howard Hughes Medical Institute investigator at UCSF and then Rockefeller University from 1995 to 2016. She was the Head of Science at the Chan Zuckerberg Initiative from 2016 to 2022. In 2012 she was awarded the $1 million Kavli Prize, and in 2013 the $3 million Breakthrough Prize in Life Sciences.

Gary Bruce Ruvkun is an American molecular biologist and Nobel laureate at Massachusetts General Hospital and professor of genetics at Harvard Medical School in Boston.

Richard Michael Durbin is a British computational biologist and Al-Kindi Professor of Genetics at the University of Cambridge. He also serves as an associate faculty member at the Wellcome Sanger Institute where he was previously a senior group leader.

sbRNA is a family of non-coding RNA first discovered in Caenorhabditis elegans. It was identified during a full transcriptome screen of the C. elegans cDNA library. Subsequent experimentation characterised sbRNA as having conserved 5' and 3' internal motifs which form a long paired stem which is interrupted with a bulge.
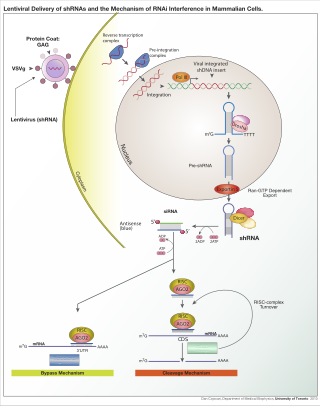
RNA interference (RNAi) is a biological process in which RNA molecules are involved in sequence-specific suppression of gene expression by double-stranded RNA, through translational or transcriptional repression. Historically, RNAi was known by other names, including co-suppression, post-transcriptional gene silencing (PTGS), and quelling. The detailed study of each of these seemingly different processes elucidated that the identity of these phenomena were all actually RNAi. Andrew Fire and Craig C. Mello shared the 2006 Nobel Prize in Physiology or Medicine for their work on RNAi in the nematode worm Caenorhabditis elegans, which they published in 1998. Since the discovery of RNAi and its regulatory potentials, it has become evident that RNAi has immense potential in suppression of desired genes. RNAi is now known as precise, efficient, stable and better than antisense therapy for gene suppression. Antisense RNA produced intracellularly by an expression vector may be developed and find utility as novel therapeutic agents.
DAF-16 is the sole ortholog of the FOXO family of transcription factors in the nematode Caenorhabditis elegans. It is responsible for activating genes involved in longevity, lipogenesis, heat shock survival and oxidative stress responses. It also protects C.elegans during food deprivation, causing it to transform into a hibernation - like state, known as a Dauer. DAF-16 is notable for being the primary transcription factor required for the profound lifespan extension observed upon mutation of the insulin-like receptor DAF-2. The gene has played a large role in research into longevity and the insulin signalling pathway as it is located in C. elegans, a successful ageing model organism.
Judith Kimble is a Henry Vilas Professor of Biochemistry, Molecular Biology, Medical Genetics and Cell and Regenerative Biology at the University of Wisconsin–Madison and Investigator with the Howard Hughes Medical Institute (HHMI). Kimble’s research focuses on the molecular regulation of animal development.

Caenorhabditis elegans- microbe interactions are defined as any interaction that encompasses the association with microbes that temporarily or permanently live in or on the nematode C. elegans. The microbes can engage in a commensal, mutualistic or pathogenic interaction with the host. These include bacterial, viral, unicellular eukaryotic, and fungal interactions. In nature C. elegans harbours a diverse set of microbes. In contrast, C. elegans strains that are cultivated in laboratories for research purposes have lost the natural associated microbial communities and are commonly maintained on a single bacterial strain, Escherichia coli OP50. However, E. coli OP50 does not allow for reverse genetic screens because RNAi libraries have only been generated in strain HT115. This limits the ability to study bacterial effects on host phenotypes. The host microbe interactions of C. elegans are closely studied because of their orthologs in humans. Therefore, the better we understand the host interactions of C. elegans the better we can understand the host interactions within the human body.
Coleen T. Murphy is a geneticist and Richard B. Fisher Preceptor in Integrative Genomics Professor of Molecular Biology at the Lewis-Sigler Institute for Integrative Genomics at Princeton University. She is director of the Paul F. Glenn Laboratories For Aging Research at Princeton.
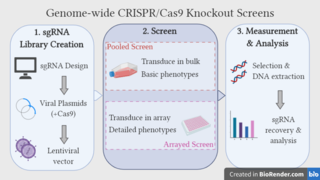
Genome-wide CRISPR-Cas9 knockout screens aim to elucidate the relationship between genotype and phenotype by ablating gene expression on a genome-wide scale and studying the resulting phenotypic alterations. The approach utilises the CRISPR-Cas9 gene editing system, coupled with libraries of single guide RNAs (sgRNAs), which are designed to target every gene in the genome. Over recent years, the genome-wide CRISPR screen has emerged as a powerful tool for performing large-scale loss-of-function screens, with low noise, high knockout efficiency and minimal off-target effects.
The Dod-13 gene in the worm Caenorhabditis elegans encoding a cytochrome p450 enzyme, which have steroid hydroxylase activity, with the CYP Symbol CYP35B1. Dod-13 is downstream gene of Daf-16 influenced the lifespan of C. elegans.

Pierre Gönczy is a Swiss and Italian cell and developmental biologist. His research focuses on centriole biology and asymmetric cell division. He is currently professor at École Polytechnique Fédérale de Lausanne (EPFL), where he directs the Laboratory of Cell and Developmental Biology.
Nathalie Le Bot is a French biologist and the chief life sciences editor of Nature Communications.












