
SR proteins are a conserved family of proteins involved in RNA splicing. SR proteins are named because they contain a protein domain with long repeats of serine and arginine amino acid residues, whose standard abbreviations are "S" and "R" respectively. SR proteins are ~200-600 amino acids in length and composed of two domains, the RNA recognition motif (RRM) region and the RS domain. SR proteins are more commonly found in the nucleus than the cytoplasm, but several SR proteins are known to shuttle between the nucleus and the cytoplasm.
RNA-binding proteins are proteins that bind to the double or single stranded RNA in cells and participate in forming ribonucleoprotein complexes. RBPs contain various structural motifs, such as RNA recognition motif (RRM), dsRNA binding domain, zinc finger and others. They are cytoplasmic and nuclear proteins. However, since most mature RNA is exported from the nucleus relatively quickly, most RBPs in the nucleus exist as complexes of protein and pre-mRNA called heterogeneous ribonucleoprotein particles (hnRNPs). RBPs have crucial roles in various cellular processes such as: cellular function, transport and localization. They especially play a major role in post-transcriptional control of RNAs, such as: splicing, polyadenylation, mRNA stabilization, mRNA localization and translation. Eukaryotic cells express diverse RBPs with unique RNA-binding activity and protein–protein interaction. According to the Eukaryotic RBP Database (EuRBPDB), there are 2961 genes encoding RBPs in humans. During evolution, the diversity of RBPs greatly increased with the increase in the number of introns. Diversity enabled eukaryotic cells to utilize RNA exons in various arrangements, giving rise to a unique RNP (ribonucleoprotein) for each RNA. Although RBPs have a crucial role in post-transcriptional regulation in gene expression, relatively few RBPs have been studied systematically.It has now become clear that RNA–RBP interactions play important roles in many biological processes among organisms.

The U4 small nuclear Ribo-Nucleic Acid is a non-coding RNA component of the major or U2-dependent spliceosome – a eukaryotic molecular machine involved in the splicing of pre-messenger RNA (pre-mRNA). It forms a duplex with U6, and with each splicing round, it is displaced from the U6 snRNA in an ATP-dependent manner, allowing U6 to re-fold and create the active site for splicing catalysis. A recycling process involving protein Brr2 releases U4 from U6, while protein Prp24 re-anneals U4 and U6. The crystal structure of a 5′ stem-loop of U4 in complex with a binding protein has been solved.

Heterogeneous nuclear ribonucleoprotein A1 is a protein that in humans is encoded by the HNRNPA1 gene. Mutations in hnRNP A1 are causative of amyotrophic lateral sclerosis and the syndrome multisystem proteinopathy.

RNA-binding protein 8A is a protein that in humans is encoded by the RBM8A gene.

Poly(rC)-binding protein 2 is a protein that in humans is encoded by the PCBP2 gene.

DEAD box proteins are involved in an assortment of metabolic processes that typically involve RNAs, but in some cases also other nucleic acids. They are highly conserved in nine motifs and can be found in most prokaryotes and eukaryotes, but not all. Many organisms, including humans, contain DEAD-box (SF2) helicases, which are involved in RNA metabolism.

Sjögren syndrome type B antigen (SS-B) also known as Lupus La protein is a protein that in humans is encoded by the SSB gene.
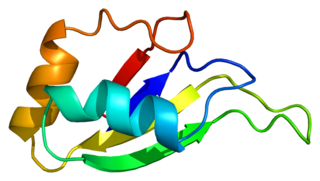
Heterogeneous nuclear ribonucleoprotein D0 (HNRNPD) also known as AU-rich element RNA-binding protein 1 (AUF1) is a protein that in humans is encoded by the HNRNPD gene. Alternative splicing of this gene results in four transcript variants.

Splicing factor 3 subunit 1 is a protein that in humans is encoded by the SF3A1 gene.
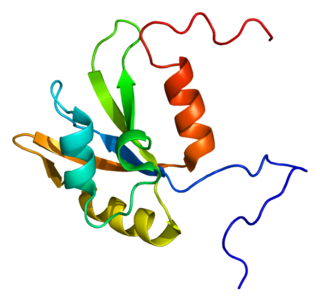
Heterogeneous nuclear ribonucleoprotein F is a protein that in humans is encoded by the HNRNPF gene.
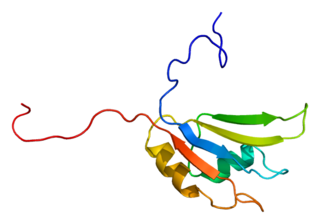
Heterogeneous nuclear ribonucleoprotein H is a protein that in humans is encoded by the HNRNPH1 gene.

Heterogeneous nuclear ribonucleoprotein L is a protein that in humans is encoded by the HNRNPL gene.

Regulator of nonsense transcripts 3B is a protein that in humans is encoded by the UPF3B gene.

Heterogeneous nuclear ribonucleoprotein D-like, also known as HNRPDL, is a protein which in humans is encoded by the HNRPDL gene.
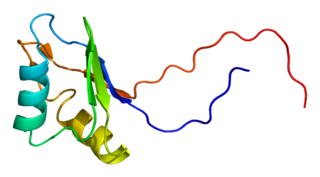
Insulin-like growth factor 2 mRNA-binding protein 2 is a protein that in humans is encoded by the IGF2BP2 gene.

Polypyrimidine tract-binding protein 1 is a protein that in humans is encoded by the PTBP1 gene.
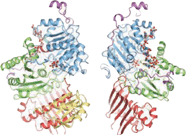
An exon junction complex (EJC) is a protein complex which forms on a pre-messenger RNA strand at the junction of two exons which have been joined together during RNA splicing. The EJC has major influences on translation, surveillance, localization of the spliced mRNA, and m6A methylation. It is first deposited onto mRNA during splicing and is then transported into the cytoplasm. There it plays a major role in post-transcriptional regulation of mRNA. It is believed that exon junction complexes provide a position-specific memory of the splicing event. The EJC consists of a stable heterotetramer core, which serves as a binding platform for other factors necessary for the mRNA pathway. The core of the EJC contains the protein eukaryotic initiation factor 4A-III bound to an adenosine triphosphate (ATP) analog, as well as the additional proteins Magoh and Y14. The binding of these proteins to nuclear speckled domains has been measured recently and it may be regulated by PI3K/AKT/mTOR signaling pathways. In order for the binding of the complex to the mRNA to occur, the eIF4AIII factor is inhibited, stopping the hydrolysis of ATP. This recognizes EJC as an ATP dependent complex. EJC also interacts with a large number of additional proteins; most notably SR proteins. These interactions are suggested to be important for mRNA compaction. The role of EJC in mRNA export is controversial.

RNA recognition motif, RNP-1 is a putative RNA-binding domain of about 90 amino acids that are known to bind single-stranded RNAs. It was found in many eukaryotic proteins.
The RNA-binding Proteins Database (RBPDB) is a biological database of RNA-binding protein specificities that includes experimental observations of RNA-binding sites. The experimental results included are both in vitro and in vivo from primary literature. It includes four metazoan species, which are Homo sapiens, Mus musculus, Drosophila melanogaster, and Caenorhabditis elegans. RNA-binding domains included in this database are RNA recognition motif, K homology, CCCH zinc finger, and more domains. As of 2021, the latest RBPDB release includes 1,171 RNA-binding proteins.


















