RNA splicing is a process in molecular biology where a newly-made precursor messenger RNA (pre-mRNA) transcript is transformed into a mature messenger RNA (mRNA). It works by removing all the introns and splicing back together exons. For nuclear-encoded genes, splicing occurs in the nucleus either during or immediately after transcription. For those eukaryotic genes that contain introns, splicing is usually needed to create an mRNA molecule that can be translated into protein. For many eukaryotic introns, splicing occurs in a series of reactions which are catalyzed by the spliceosome, a complex of small nuclear ribonucleoproteins (snRNPs). There exist self-splicing introns, that is, ribozymes that can catalyze their own excision from their parent RNA molecule. The process of transcription, splicing and translation is called gene expression, the central dogma of molecular biology.

A spliceosome is a large ribonucleoprotein (RNP) complex found primarily within the nucleus of eukaryotic cells. The spliceosome is assembled from small nuclear RNAs (snRNA) and numerous proteins. Small nuclear RNA (snRNA) molecules bind to specific proteins to form a small nuclear ribonucleoprotein complex, which in turn combines with other snRNPs to form a large ribonucleoprotein complex called a spliceosome. The spliceosome removes introns from a transcribed pre-mRNA, a type of primary transcript. This process is generally referred to as splicing. An analogy is a film editor, who selectively cuts out irrelevant or incorrect material from the initial film and sends the cleaned-up version to the director for the final cut.

SR proteins are a conserved family of proteins involved in RNA splicing. SR proteins are named because they contain a protein domain with long repeats of serine and arginine amino acid residues, whose standard abbreviations are "S" and "R" respectively. SR proteins are ~200-600 amino acids in length and composed of two domains, the RNA recognition motif (RRM) region and the RS domain. SR proteins are more commonly found in the nucleus than the cytoplasm, but several SR proteins are known to shuttle between the nucleus and the cytoplasm.

Nucleoproteins are proteins conjugated with nucleic acids. Typical nucleoproteins include ribosomes, nucleosomes and viral nucleocapsid proteins.
RNA-binding proteins are proteins that bind to the double or single stranded RNA in cells and participate in forming ribonucleoprotein complexes. RBPs contain various structural motifs, such as RNA recognition motif (RRM), dsRNA binding domain, zinc finger and others. They are cytoplasmic and nuclear proteins. However, since most mature RNA is exported from the nucleus relatively quickly, most RBPs in the nucleus exist as complexes of protein and pre-mRNA called heterogeneous ribonucleoprotein particles (hnRNPs). RBPs have crucial roles in various cellular processes such as: cellular function, transport and localization. They especially play a major role in post-transcriptional control of RNAs, such as: splicing, polyadenylation, mRNA stabilization, mRNA localization and translation. Eukaryotic cells express diverse RBPs with unique RNA-binding activity and protein–protein interaction. According to the Eukaryotic RBP Database (EuRBPDB), there are 2961 genes encoding RBPs in humans. During evolution, the diversity of RBPs greatly increased with the increase in the number of introns. Diversity enabled eukaryotic cells to utilize RNA exons in various arrangements, giving rise to a unique RNP (ribonucleoprotein) for each RNA. Although RBPs have a crucial role in post-transcriptional regulation in gene expression, relatively few RBPs have been studied systematically.It has now become clear that RNA–RBP interactions play important roles in many biological processes among organisms.
Small nuclear RNA (snRNA) is a class of small RNA molecules that are found within the splicing speckles and Cajal bodies of the cell nucleus in eukaryotic cells. The length of an average snRNA is approximately 150 nucleotides. They are transcribed by either RNA polymerase II or RNA polymerase III. Their primary function is in the processing of pre-messenger RNA (hnRNA) in the nucleus. They have also been shown to aid in the regulation of transcription factors or RNA polymerase II, and maintaining the telomeres.

In molecular biology, LSm proteins are a family of RNA-binding proteins found in virtually every cellular organism. LSm is a contraction of 'like Sm', because the first identified members of the LSm protein family were the Sm proteins. LSm proteins are defined by a characteristic three-dimensional structure and their assembly into rings of six or seven individual LSm protein molecules, and play a large number of various roles in mRNA processing and regulation.
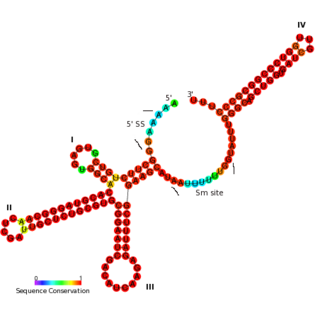
The U11 snRNA is an important non-coding RNA in the minor spliceosome protein complex, which activates the alternative splicing mechanism. The minor spliceosome is associated with similar protein components as the major spliceosome. It uses U11 snRNA to recognize the 5' splice site while U12 snRNA binds to the branchpoint to recognize the 3' splice site.

U1 spliceosomal RNA is the small nuclear RNA (snRNA) component of U1 snRNP, an RNA-protein complex that combines with other snRNPs, unmodified pre-mRNA, and various other proteins to assemble a spliceosome, a large RNA-protein molecular complex upon which splicing of pre-mRNA occurs. Splicing, or the removal of introns, is a major aspect of post-transcriptional modification, and takes place only in the nucleus of eukaryotes.

The U4 small nuclear Ribo-Nucleic Acid is a non-coding RNA component of the major or U2-dependent spliceosome – a eukaryotic molecular machine involved in the splicing of pre-messenger RNA (pre-mRNA). It forms a duplex with U6, and with each splicing round, it is displaced from the U6 snRNA in an ATP-dependent manner, allowing U6 to re-fold and create the active site for splicing catalysis. A recycling process involving protein Brr2 releases U4 from U6, while protein Prp24 re-anneals U4 and U6. The crystal structure of a 5′ stem-loop of U4 in complex with a binding protein has been solved.

rRNA 2'-O-methyltransferase fibrillarin is an enzyme that in humans is encoded by the FBL gene.

Splicing factor U2AF 65 kDa subunit is a protein that in humans is encoded by the U2AF2 gene.

snRNP70 also known as U1 small nuclear ribonucleoprotein 70 kDa is a protein that in humans is encoded by the SNRNP70 gene. snRNP70 is a small nuclear ribonucleoprotein that associates with U1 spliceosomal RNA, forming the U1snRNP a core component of the spliceosome. The U1-70K protein and other components of the spliceosome complex form detergent-insoluble aggregates in both sporadic and familial human cases of Alzheimer's disease. U1-70K co-localizes with Tau in neurofibrillary tangles in Alzheimer's disease.

DEAD box proteins are involved in an assortment of metabolic processes that typically involve RNAs, but in some cases also other nucleic acids. They are highly conserved in nine motifs and can be found in most prokaryotes and eukaryotes, but not all. Many organisms, including humans, contain DEAD-box (SF2) helicases, which are involved in RNA metabolism.

Small nuclear ribonucleoprotein-associated proteins B and B' is a protein that in humans is encoded by the SNRPB gene.

U1 small nuclear ribonucleoprotein A is a protein that in humans is encoded by the SNRPA gene.
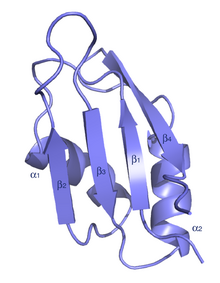
U1 small nuclear ribonucleoprotein C is a protein that in humans is encoded by the SNRPC gene.

Prp24 is a protein part of the pre-messenger RNA splicing process and aids the binding of U6 snRNA to U4 snRNA during the formation of spliceosomes. Found in eukaryotes from yeast to E. coli, fungi, and humans, Prp24 was initially discovered to be an important element of RNA splicing in 1989. Mutations in Prp24 were later discovered in 1991 to suppress mutations in U4 that resulted in cold-sensitive strains of yeast, indicating its involvement in the reformation of the U4/U6 duplex after the catalytic steps of splicing.

In molecular biology, the La domain is a conserved protein domain. Human 60 kDa SS-A/Ro ribonucleoproteins (RNPs) are composed of one of the four small Y RNAs and at least two proteins, Ro60 and La. The La protein is a 47 kDa polypeptide that frequently acts as an autoantigen in systemic lupus erythematosus and Sjögren syndrome. In the nucleus, La acts as a RNA polymerase III transcription factor, while in the cytoplasm, La acts as a translation factor. In the nucleus, La binds to the 3'UTR of nascent RNAP III transcripts to assist in folding and maturation. In the cytoplasm, La recognises specific classes of mRNAs that contain a 5'-terminal oligopyrimidine (5'TOP) motif known to control protein synthesis. The specific recognition is mediated by the N-terminal domain of La, which comprises a La motif and an RNA recognition motif (RRM). The La motif adopts an alpha/beta fold that comprises a winged-helix motif.

Kiyoshi Nagai was a Japanese structural biologist at the MRC Laboratory of Molecular Biology Cambridge, UK. He was known for his work on the mechanism of RNA splicing and structures of the spliceosome.