Structure and function
This gene belongs to the subfamily of ubiquitously expressed heterogeneous nuclear ribonucleoproteins (hnRNPs). The hnRNPs are RNA-binding proteins and they complex with heterogeneous nuclear RNA (hnRNA).
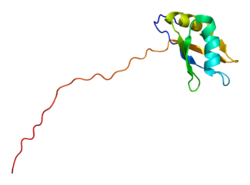
SYNCRIP is made up of an N-terminal helix bundle known as the “acidic domain” (AcD), followed by three sequential RNA recognition motifs (RRMs) separated by short linkers, and an arginine-glycine-rich domain called the "RGG box" at the C-terminus. The RRMs play a role in RNA binding, while the AcD engages in protein-protein interactions (PPIs). The RGG box is involved in both RNA binding and in PPIs. The AcD is unique to SYNCRIP and its nuclear homolog hnRNP R, and is involved in interactions with APOBEC-1. [11] It is a self-folding, all-helical domain of five α-helices, containing a large hydrophobic cavity and a positively charged surface area as potential interaction sites in addition to negatively charged surface areas, with no structural homologs in any other known proteins. [11] [12] The hydrophobic core is mostly made up of leucine residues, while the surface is made up of 15 acidic residues and 13 basic residues which together form a vast interaction network. [12] The AcD is linked to RRM1 by a unique α-β-β unit, creating an “extended RRM fold” mediated primarily by hydrophobic interactions. [12] The RGG box is an unstructured region containing numerous Arg-Gly-Gly repeats with fairly regular spacing. There are eight such repeats in SYNCRIP. This domain can bind proteins and RNA independently, even if the other binding domains are not present. [13] Although this domain is rich in arginine content, it does not have any arginine-rich clusters as might be observed in usual arginine-rich RBPs. [13]
Isoform 1 is a component of the apolipoprotein B (apoB) mRNA editosome complex, and it modulates the post-transcriptional C-to-U RNA editing of apoB mRNA through binding either to the apoB mRNA-editing enzyme catalytic peptide 1 (APOBEC-1), to the APOBEC-1 complementation factor (ACF), or directly to RNA itself. [6] Isoform 1 is also implicated with other RBPs in the cytoplasmic de-adenylation and translational and decay interplay of c-Fos mRNA mediated by the major coding-region determinant of instability (mCRD) domain. [14]
The function of isoform 2 is not as clearly understood.
Isoform 3 is involved in cytoplasmic vesicle-based mRNA transport through interaction with synaptotagmins (SYTs). [8] This isoform is also a component of the gamma interferon (IFNγ)-activated inhibitor of translation (GAIT) complex in humans, which mediates IFNγ-induced transcript-selective translation inhibition in inflammation processes. [15] Upon IFN-γ activation, SYNCRIP assembles into the GAIT complex, which binds to stem-loop-containing GAIT elements in the 3’-untranslated region (3’- UTR) of diverse inflammatory mRNAs and suppresses their translation, but this seems to not be essential for the overall function of the GAIT complex. [15]
This page is based on this
Wikipedia article Text is available under the
CC BY-SA 4.0 license; additional terms may apply.
Images, videos and audio are available under their respective licenses.