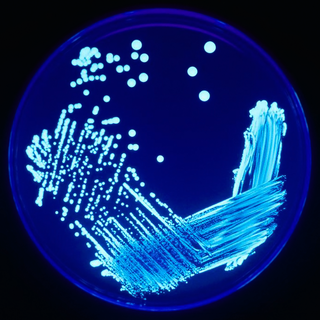
Legionella is a genus of pathogenic gram-negative bacteria that includes the species L. pneumophila, causing legionellosis including a pneumonia-type illness called Legionnaires' disease and a mild flu-like illness called Pontiac fever.
Atypical pneumonia, also known as walking pneumonia, is any type of pneumonia not caused by one of the pathogens most commonly associated with the disease. Its clinical presentation contrasts to that of "typical" pneumonia. A variety of microorganisms can cause it. When it develops independently from another disease, it is called primary atypical pneumonia (PAP).

Legionella pneumophila is a thin, aerobic, pleomorphic, flagellated, non-spore-forming, Gram-negative bacterium of the genus Legionella. L. pneumophila is the primary human pathogenic bacterium in this group and is the causative agent of Legionnaires' disease, also known as legionellosis.
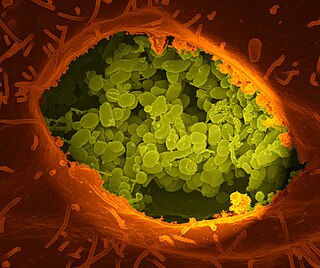
Coxiella burnetii is an obligate intracellular bacterial pathogen, and is the causative agent of Q fever. The genus Coxiella is morphologically similar to Rickettsia, but with a variety of genetic and physiological differences. C. burnetii is a small Gram-negative, coccobacillary bacterium that is highly resistant to environmental stresses such as high temperature, osmotic pressure, and ultraviolet light. These characteristics are attributed to a small cell variant form of the organism that is part of a biphasic developmental cycle, including a more metabolically and replicatively active large cell variant form. It can survive standard disinfectants, and is resistant to many other environmental changes like those presented in the phagolysosome.

The Legionellales are an order of Pseudomonadota. Like all Pseudomonadota, they are Gram-negative. They comprise two families, typified by Legionella and Coxiella, both of which include notable pathogens. For example, Q fever is caused by Coxiella burnetii and Legionella pneumophila causes Legionnaires' disease and Pontiac fever.

Staphylococcus epidermidis is a Gram-positive bacterium, and one of over 40 species belonging to the genus Staphylococcus. It is part of the normal human microbiota, typically the skin microbiota, and less commonly the mucosal microbiota and also found in marine sponges. It is a facultative anaerobic bacteria. Although S. epidermidis is not usually pathogenic, patients with compromised immune systems are at risk of developing infection. These infections are generally hospital-acquired. S. epidermidis is a particular concern for people with catheters or other surgical implants because it is known to form biofilms that grow on these devices. Being part of the normal skin microbiota, S. epidermidis is a frequent contaminant of specimens sent to the diagnostic laboratory.
Intracellular parasites are microparasites that are capable of growing and reproducing inside the cells of a host.

Xanthomonas campestris is a gram-negative, obligate aerobic bacterium that is a member of the Xanthomonas genus, which is a group of bacteria that are commonly known for their association with plant disease. The species is considered to be dominant amongst its genus, as it originally had over 140 identified pathovars and has been found to infect both monocotyledonous and dicotyledonous plants of economical value with various plant diseases. This includes "black rot" in cruciferous vegetables, bacterial wilt of turfgrass, bacterial blight, and leaf spot, for example.
Dictyostelium discoideum is a species of soil-dwelling amoeba belonging to the phylum Amoebozoa, infraphylum Mycetozoa. Commonly referred to as slime mold, D. discoideum is a eukaryote that transitions from a collection of unicellular amoebae into a multicellular slug and then into a fruiting body within its lifetime. Its unique asexual life cycle consists of four stages: vegetative, aggregation, migration, and culmination. The life cycle of D. discoideum is relatively short, which allows for timely viewing of all stages. The cells involved in the life cycle undergo movement, chemical signaling, and development, which are applicable to human cancer research. The simplicity of its life cycle makes D. discoideum a valuable model organism to study genetic, cellular, and biochemical processes in other organisms.

Pathogenic bacteria are bacteria that can cause disease. This article focuses on the bacteria that are pathogenic to humans. Most species of bacteria are harmless and are often beneficial but others can cause infectious diseases. The number of these pathogenic species in humans is estimated to be fewer than a hundred. By contrast, several thousand species are part of the gut flora present in the digestive tract.
Willaertia /ˈwɪləɹʃə/ is a genus of non-pathogenic, free-living, thermophilic amoebae in the family Vahlkampfiidae.
Legionella micdadei is a Gram-negative bacterium from the genus Legionella, which stains acid-fast. It stains weakly, but loses this trait upon being grown in culture. Tatlockia micdadei is an alternative term for L. micdadei, the Pittsburgh pneumonia agent and TATLOCK strain. It is named after Joseph E. McDade, who first isolated L. pneumophila.

The 1976 Legionnaires disease outbreak, occurring in the late summer in Philadelphia, Pennsylvania, United States was the first occasion in which a cluster of a particular type of pneumonia cases were determined to be caused by the Legionella pneumophila bacteria.
Variovorax paradoxus is a gram negative, beta proteobacterium from the genus Variovorax. Strains of V. paradoxus can be categorized into two groups, hydrogen oxidizers and heterotrophic strains, both of which are aerobic. The genus name Vario-vorax and species name para-doxus (contrary-opinion) reflects both the dichotomy of V. paradoxus metabolisms, but also its ability to utilize a wide array of organic compounds.
Legionella anisa is a Gram-negative bacterium, one of more than 40 species in the family Legionellaceae. After Legionella pneumophila, this species has been isolated most frequently from water samples. This species is also one of the several pathogenic forms of Legionella having been associated with rare clinical cases of illness including Pontiac fever and Legionnaires' disease.
Legionella cherrii is an aerobic, flagellated, Gram-negative bacterium from the genus Legionella. It was isolated from a heated water sample in Minnesota. L. cherrii is similar to another Legionella species, L. pneumophila, and is believed to cause major respiratory problems.
Legionella jordanis is a Gram-negative bacterium from the genus Legionella which was isolated from the Jordan River in Bloomington, Indiana and from the sewage in DeKalb County, Georgia. L. jordanis is a rare human pathogen and can cause respiratory tract infections.

Legionnaires' disease is a form of atypical pneumonia caused by any species of Legionella bacteria, quite often Legionella pneumophila. Signs and symptoms include cough, shortness of breath, high fever, muscle pains, and headaches. Nausea, vomiting, and diarrhea may also occur. This often begins 2–10 days after exposure.
Proteobiotics are natural metabolites which are produced by fermentation process of specific probiotic strains. These small oligopeptides were originally discovered in and isolated from culture media used to grow probiotic bacteria and may account for some of the health benefits of probiotics.

Amoebozoa of the free living genus Acanthamoeba and the social amoeba genus Dictyostelium are single celled eukaryotic organisms that feed on bacteria, fungi, and algae through phagocytosis, with digestion occurring in phagolysosomes. Amoebozoa are present in most terrestrial ecosystems including soil and freshwater. Amoebozoa contain a vast array of symbionts that range from transient to permanent infections, confer a range of effects from mutualistic to pathogenic, and can act as environmental reservoirs for animal pathogenic bacteria. As single celled phagocytic organisms, amoebas simulate the function and environment of immune cells like macrophages, and as such their interactions with bacteria and other microbes are of great importance in understanding functions of the human immune system, as well as understanding how microbiomes can originate in eukaryotic organisms.









