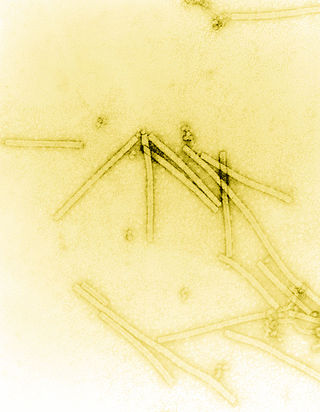
Tobacco mosaic virus (TMV) is a positive-sense single-stranded RNA virus species in the genus Tobamovirus that infects a wide range of plants, especially tobacco and other members of the family Solanaceae. The infection causes characteristic patterns, such as "mosaic"-like mottling and discoloration on the leaves. TMV was the first virus to be discovered. Although it was known from the late 19th century that a non-bacterial infectious disease was damaging tobacco crops, it was not until 1930 that the infectious agent was determined to be a virus. It is the first pathogen identified as a virus. The virus was crystallised by W.M. Stanley. It has a similar size to the largest synthetic molecule, known as PG5.

Rubella virus (RuV) is the pathogenic agent of the disease rubella, transmitted only between humans via the respiratory route, and is the main cause of congenital rubella syndrome when infection occurs during the first weeks of pregnancy.

Plant viruses are viruses that affect plants. Like all other viruses, plant viruses are obligate intracellular parasites that do not have the molecular machinery to replicate without a host. Plant viruses can be pathogenic to vascular plants.

Potyviridae is a family of positive-strand RNA viruses that encompasses more than 30% of known plant viruses, many of which are of great agricultural significance. The family has 12 genera and 235 species, three of which are unassigned to a genus.

Tombusviridae is a family of single-stranded positive sense RNA plant viruses. There are three subfamilies, 17 genera, and 95 species in this family. The name is derived from Tomato bushy stunt virus (TBSV).

Nidovirales is an order of enveloped, positive-strand RNA viruses which infect vertebrates and invertebrates. Host organisms include mammals, birds, reptiles, amphibians, fish, arthropods, molluscs, and helminths. The order includes the families Coronaviridae, Arteriviridae, Roniviridae, and Mesoniviridae.
Ribosome shunting is a mechanism of translation initiation in which ribosomes bypass, or "shunt over", parts of the 5' untranslated region to reach the start codon. However, a benefit of ribosomal shunting is that it can translate backwards allowing more information to be stored than usual in an mRNA molecule. Some viral RNAs have been shown to use ribosome shunting as a more efficient form of translation during certain stages of viral life cycle or when translation initiation factors are scarce. Some viruses known to use this mechanism include adenovirus, Sendai virus, human papillomavirus, duck hepatitis B pararetrovirus, rice tungro bacilliform viruses, and cauliflower mosaic virus. In these viruses the ribosome is directly translocated from the upstream initiation complex to the start codon (AUG) without the need to unwind RNA secondary structures.

Tomato bushy stunt virus (TBSV) is a virus of the tombusvirus family. It was first reported in tomatoes in 1935 and primarily affects vegetable crops, though it is not generally considered an economically significant plant pathogen. Depending upon the host, TBSV causes stunting of growth, leaf mottling, and deformed or absent fruit. The virus is likely to be soil-borne in the natural setting, but can also transmitted mechanically, for example through contaminated cutting tools. TBSV has been used as a model system in virology research on the life cycle of plant viruses, particularly in experimental infections of the model host plant Nicotiana benthamiana.
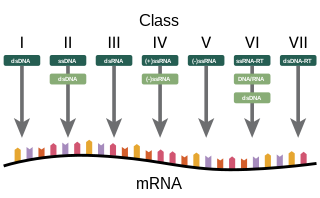
Baltimore classification is a system used to classify viruses based on their manner of messenger RNA (mRNA) synthesis. By organizing viruses based on their manner of mRNA production, it is possible to study viruses that behave similarly as a distinct group. Seven Baltimore groups are described that take into consideration whether the viral genome is made of deoxyribonucleic acid (DNA) or ribonucleic acid (RNA), whether the genome is single- or double-stranded, and whether the sense of a single-stranded RNA genome is positive or negative.
Drosophila X virus (DXV) belongs to the Birnaviridae family of viruses. Birnaviridae currently consists of three genera. The first genus is Entomobirnavirus, which contains DXV. The next genus is Aquabirnavirus, containing infectious pancreatic necrosis virus (IPNV). The last genus is Avibirnavirus, which contains infectious bursal disease virus (IBDV). All of these genera contain homology in three specific areas of their transcripts. The homology comes from the amino and carboxyl regions of preVP2, a small 21-residue-long domain near the carboxyl terminal of VP3, and similar small ORFs sequences.

Deformed wing virus (DWV) is an RNA virus, one of 22 known viruses affecting honey bees. While most commonly infecting the honey bee, Apis mellifera, it has also been documented in other bee species, like Bombus terrestris, thus, indicating it may have a wider host specificity than previously anticipated. The virus was first isolated from a sample of symptomatic honeybees from Japan in the early 1980s and is currently distributed worldwide. It is found also in pollen baskets and commercially reared bumblebees. Its main vector in A. mellifera is the Varroa mite. It is named after what is usually the most obvious deformity it induces in the development of a honeybee pupa, which is shrunken and deformed wings, but other developmental deformities are often present.

Partitiviridae is a family of double-stranded RNA viruses. Plants, fungi, and protozoa serve as natural hosts. It has been suggested that they can also infect bacteria. The name comes from the Latin partitius, which means divided, and refers to the segmented genome of partitiviruses. There are five genera and 60 species in the family, 15 of which are unassigned to a genus.

Alfalfa mosaic virus (AMV), also known as Lucerne mosaic virus or Potato calico virus, is a worldwide distributed phytopathogen that can lead to necrosis and yellow mosaics on a large variety of plant species, including commercially important crops. It is the only Alfamovirus of the family Bromoviridae. In 1931 Weimer J.L. was the first to report AMV in alfalfa. Transmission of the virus occurs mainly by some aphids, by seeds or by pollen to the seed.
Idaeovirus is a genus of positive-sense ssRNA viruses that contains two species: Raspberry bushy dwarf virus (RBDV) and Privet idaeovirus. RBDV has two host-dependent clades: one for raspberries; the other for grapevines. Infections are a significant agricultural burden, resulting in decreased yield and quality of crops. RBDV has a synergistic relation with Raspberry leaf mottle virus, with co-infection greatly amplifying the concentration of virions in infected plants. The virus is transmitted via pollination with RBDV-infected pollen grains that first infect the stigma before causing systemic infection.

Potexvirus is a genus of pathogenic viruses in the order Tymovirales, in the family Alphaflexiviridae. Plants serve as natural hosts. There are 48 species in this genus, three of which are assigned to a subgenus. Diseases associated with this genus include: mosaic and ringspot symptoms. The genus name comes from POTato virus X).

Carlavirus, formerly known as the "Carnation latent virus group", is a genus of viruses in the order Tymovirales, in the family Betaflexiviridae. Plants serve as natural hosts. There are 53 species in this genus. Diseases associated with this genus include: mosaic and ringspot symptoms.
Donkey orchid symptomless virus (DOSV) was first discovered in Perth, Australia which is located on the Western side of Australia. The Southwest Australian Floristic Region is very biodiverse and secluded with deserts on the east and northeast side along with the ocean on the south side. It is home to approximately 8,000 indigenous plant species that are accustomed to living in a Mediterranean climate. This type of climate means that the plants have adapted to living in unfertile soil, dryness, and can withstand frequent fires in the area. Out of 394 terrestrial orchid species, 76 of these species are threatened. Reasons contributing to plants being threatened in this area could be due to the rate at which they reproduce fruit, pollinate, or human contact.
ORF3b is a gene found in coronaviruses of the subgenus Sarbecovirus, encoding a short non-structural protein. It is present in both SARS-CoV and SARS-CoV-2, though the protein product has very different lengths in the two viruses. The encoded protein is significantly shorter in SARS-CoV-2, at only 22 amino acid residues compared to 153-155 in SARS-CoV. Both the longer SARS-CoV and shorter SARS-CoV-2 proteins have been reported as interferon antagonists. It is unclear whether the SARS-CoV-2 gene expresses a functional protein.
ORF1ab refers collectively to two open reading frames (ORFs), ORF1a and ORF1b, that are conserved in the genomes of nidoviruses, a group of viruses that includes coronaviruses. The genes express large polyproteins that undergo proteolysis to form several nonstructural proteins with various functions in the viral life cycle, including proteases and the components of the replicase-transcriptase complex (RTC). Together the two ORFs are sometimes referred to as the replicase gene. They are related by a programmed ribosomal frameshift that allows the ribosome to continue translating past the stop codon at the end of ORF1a, in a -1 reading frame. The resulting polyproteins are known as pp1a and pp1ab.















