
Listeria monocytogenes is the species of pathogenic bacteria that causes the infection listeriosis. It is a facultative anaerobic bacterium, capable of surviving in the presence or absence of oxygen. It can grow and reproduce inside the host's cells and is one of the most virulent foodborne pathogens, with 20 to 30% of foodborne listeriosis infections in high-risk individuals may be fatal. Responsible for an estimated 1,600 illnesses and 260 deaths in the United States annually, listeriosis ranks third in total number of deaths among foodborne bacterial pathogens, with fatality rates exceeding even Salmonella spp. and Clostridium botulinum. In the European Union, listeriosis follows an upward trend that began in 2008, causing 2,161 confirmed cases and 210 reported deaths in 2014, 16% more than in 2013. Listeriosis mortality rates are also higher in the EU than for other foodborne pathogens.

Shigella flexneri is a species of Gram-negative bacteria in the genus Shigella that can cause diarrhea in humans. Several different serogroups of Shigella are described; S. flexneri belongs to group B. S. flexneri infections can usually be treated with antibiotics, although some strains have become resistant. Less severe cases are not usually treated because they become more resistant in the future.
The gene rpoS encodes the sigma factor sigma-38, a 37.8 kD protein in Escherichia coli. Sigma factors are proteins that regulate transcription in bacteria. Sigma factors can be activated in response to different environmental conditions. rpoS is transcribed in late exponential phase, and RpoS is the primary regulator of stationary phase genes. RpoS is a central regulator of the general stress response and operates in both a retroactive and a proactive manner: it not only allows the cell to survive environmental challenges, but it also prepares the cell for subsequent stresses (cross-protection). The transcriptional regulator CsgD is central to biofilm formation, controlling the expression of the curli structural and export proteins, and the diguanylate cyclase, adrA, which indirectly activates cellulose production. The rpoS gene most likely originated in the gammaproteobacteria.

The gcvB RNA gene encodes a small non-coding RNA involved in the regulation of a number of amino acid transport systems as well as amino acid biosynthetic genes. The GcvB gene is found in enteric bacteria such as Escherichia coli. GcvB regulates genes by acting as an antisense binding partner of the mRNAs for each regulated gene. This binding is dependent on binding to a protein called Hfq. Transcription of the GcvB RNA is activated by the adjacent GcvA gene and repressed by the GcvR gene. A deletion of GcvB RNA from Y. pestis changed colony shape as well as reducing growth. It has been shown by gene deletion that GcvB is a regulator of acid resistance in E. coli. GcvB enhances the ability of the bacterium to survive low pH by upregulating the levels of the alternate sigma factor RpoS. A polymeric form of GcvB has recently been identified. Interaction of GcvB with small RNA SroC triggers the degradation of GcvB by RNase E, lifting the GcvB-mediated mRNA repression of its target genes.

The OmrA-B RNA gene family is a pair of homologous OmpR-regulated small non-coding RNA that was discovered in E. coli during two large-scale screens. OmrA-B is highly abundant in stationary phase, but low levels could be detected in exponentially growing cells as well. RygB is adjacent to RygA a closely related RNA. These RNAs bind to the Hfq protein and regulate gene expression by antisense binding. They negatively regulate the expression of several genes encoding outer membrane proteins, including cirA, CsgD, fecA, fepA and ompT by binding in the vicinity of the Shine-Dalgarno sequence, suggesting the control of these targets is dependent on Hfq protein and RNase E. Taken together, these data suggest that OmrA-B participates in the regulation of outer membrane composition, responding to environmental conditions.
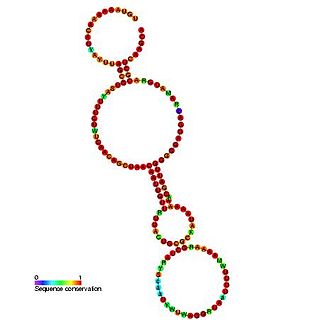
The PrfA thermoregulator UTR is an RNA thermometer found in the 5' UTR of the prfA gene. In Listeria monocytogenes, virulence genes are maximally expressed at 37 °C but are almost silent at 30 °C. The genes are controlled by PrfA, a transcriptional activator whose expression is thermoregulated. It has been shown that the untranslated mRNA (UTR) preceding prfA, forms a secondary structure, which masks the ribosome binding region. It is thought that at 37 °C, the hairpin structure 'melts' and the SD sequence is unmasked.

In molecular biology the ArcZ RNA is a small non-coding RNA (ncRNA). It is the functional product of a gene which is not translated into protein. ArcZ is an Hfq binding RNA that functions as an antisense regulator of a number of protein coding genes.
Listeriolysin O (LLO) is a hemolysin produced by the bacterium Listeria monocytogenes, the pathogen responsible for causing listeriosis. The toxin may be considered a virulence factor, since it is crucial for the virulence of L. monocytogenes.

Listeria Hfq binding LhrA is a ncRNA that was identified by screening for RNA molecules which co-immunoprecipitated with the RNA chaperone Hfq. This RNA is transcribed from a region overlapping with a predicted protein of unknown function (Lmo2257) and is located between a putative intracellular protease and a putative protein of the ribulose-phosphate 3 epimerase family. It is highly expressed in the stationary growth phase but the function is unknown. It is proposed to be a regulatory RNA which controls gene expression at the post transcriptional level by binding the target mRNA in an Hfq dependent fashion. This RNA molecule appears to be conserved amongst Listeria species but has not been identified in other bacterial species.
Internalins are surface proteins found on Listeria monocytogenes. They exist in two known forms, InlA and InlB. They are used by the bacteria to invade mammalian cells via cadherins transmembrane proteins and Met receptors respectively. The exact role of these proteins and their invasiveness in vivo is not completely understood. In cultured cells, InlA is necessary to facilitate Listeria entry into human epithelial cells, while InlB is necessary for Listeria internalisation in several other cell types, including hepatocytes, fibroblasts, and epithelioid cells. Internalins are mainly surface-exposed virulence factors present in a number of Gram-positive bacteria whose role ranges from recognition of cellular receptors to aid in pathogen entry to escape from autophagy.

An Hfq binding sRNA is an sRNA that binds the bacterial RNA binding protein called Hfq. A number of bacterial small RNAs which have been shown to bind to Hfq have been characterised . Many of these RNAs share a similar structure composed of three stem-loops. Several studies have expanded this list, and experimentally validated a total of 64 Hfq binding sRNA in Salmonella Typhimurium. A transcriptome wide study on Hfq binding sites in Salmonella mapped 126 Hfq binding sites within sRNAs. Genomic SELEX has been used to show that Hfq binding RNAs are enriched in the sequence motif 5′-AAYAAYAA-3′. Genome-wide study identified 40 candidate Hfq-dependent sRNAs in plant pathogen Erwinia amylovora. 12 of them were confirmed by Northern blot.

Listeria monocytogenes is a gram positive bacterium and causes many food-borne infections such as Listeriosis. This bacteria is ubiquitous in the environment where it can act as either a saprophyte when free living within the environment or as a pathogen when entering a host organism. Many non-coding RNAs have been identified within the bacteria genome where several of these have been classified as novel non-coding RNAs and may contribute to pathogenesis.

The c4 antisense RNA is a non-coding RNA used by certain phages that infect bacteria. It was initially identified in the P1 and P7 phages of E. coli. The identification of c4 antisense RNAs solved the mystery of the mechanism for regulation of the ant gene, which is an anti-repressor.

The Actin assembly-inducing protein (ActA) is a protein encoded and used by Listeria monocytogenes to propel itself through a mammalian host cell. ActA is a bacterial surface protein comprising a membrane-spanning region. In a mammalian cell the bacterial ActA interacts with the Arp2/3 complex and actin monomers to induce actin polymerization on the bacterial surface generating an actin comet tail. The gene encoding ActA is named actA or prtB.

FourU thermometers are a class of non-coding RNA thermometers found in Salmonella. They are named 'FourU' due to the four highly conserved uridine nucleotides found directly opposite the Shine-Dalgarno sequence on hairpin II (pictured). RNA thermometers such as FourU control regulation of temperature via heat shock proteins in many prokaryotes. FourU thermometers are relatively small RNA molecules, only 57 nucleotides in length, and have a simple two-hairpin structure.
Rsa RNAs are non-coding RNAs found in the bacterium Staphylococcus aureus. The shared name comes from their discovery, and does not imply homology. Bioinformatics scans identified the 16 Rsa RNA families named RsaA-K and RsaOA-OG. Others, RsaOH-OX, were found thanks to an RNomic approach. Although the RNAs showed varying expression patterns, many of the newly discovered RNAs were shown to be Hfq-independent and most carried a C-rich motif (UCCC).
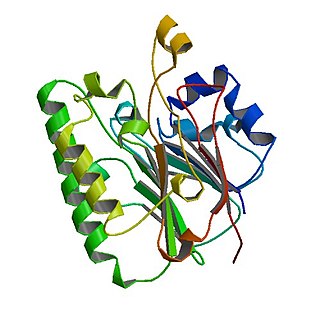
The Catabolite repression control (Crc) protein participates in suppressing expression of several genes involved in utilization of carbon sources in Pseudomonas bacteria. Presence of organic acids triggers activation of Crc and in conjunction with the Hfq protein genes that metabolize a given carbon source are downregulated until another more favorable carbon source is depleted. Crc-mediated regulation impact processes such as biofilm formation, virulence and antibiotic susceptibility.
Bacterial small RNAs (sRNA) are an important class of regulatory molecules in bacteria such as Brucella. They are often bound to the chaperone protein Hfq, which allows them to interact with mRNA(s). In Brucella suis 1330 RNA sequencing identified a novel list of 33 sRNAs and 62 Hfq-associated mRNAs. In Brucella melitensis eight novel sRNA genes were identified using bioinformatic and experimental approach. One of them BSR0602 was found to modulate the intracellular survival of B. melitensis. In another large-scale deep sequencing study 1321 sRNAs were identified in B. melitensis. BSR0441 sRNA was further investigated in this study and shown to play role in the intracellular survival. sRNA BM-sr0117 from Brucella melitensis was identified and shown to be bound to and cleaved by Bm-RNase III. AbcR and AbcR2 were studied B. abortus. Seven novel sRNAs were validated and their interaction with a putative target sequence was verified in B. abortus.
The SraL RNA, also known as RyjA, is a small non-coding RNA discovered in E. coli, and later in Salmonella Tiphimurium. This ncRNA was found to be expressed only in stationary phase. It may possibly play a role in Salmonella virulence. The major stationary phase regulator RpoS is transcriptionally regulating SraL and directly binds to the sraL gene promoter. SraL down-regulates the expression of the ribosome-associated chaperone Trigger Factor (TF), which is involved in the folding of the newly synthesised cystolic proteins.















