
Tumor necrosis factor is an adipokine and a cytokine. TNF is a member of the TNF superfamily, which consists of various transmembrane proteins with a homologous TNF domain.

The insulin receptor (IR) is a transmembrane receptor that is activated by insulin, IGF-I, IGF-II and belongs to the large class of receptor tyrosine kinase. Metabolically, the insulin receptor plays a key role in the regulation of glucose homeostasis; a functional process that under degenerate conditions may result in a range of clinical manifestations including diabetes and cancer. Insulin signalling controls access to blood glucose in body cells. When insulin falls, especially in those with high insulin sensitivity, body cells begin only to have access to lipids that do not require transport across the membrane. So, in this way, insulin is the key regulator of fat metabolism as well. Biochemically, the insulin receptor is encoded by a single gene INSR, from which alternate splicing during transcription results in either IR-A or IR-B isoforms. Downstream post-translational events of either isoform result in the formation of a proteolytically cleaved α and β subunit, which upon combination are ultimately capable of homo or hetero-dimerisation to produce the ≈320 kDa disulfide-linked transmembrane insulin receptor.
Members of the signal transducer and activator of transcription (STAT) protein family are intracellular transcription factors that mediate many aspects of cellular immunity, proliferation, apoptosis and differentiation. They are primarily activated by membrane receptor-associated Janus kinases (JAK). Dysregulation of this pathway is frequently observed in primary tumors and leads to increased angiogenesis which enhances the survival of tumors and immunosuppression. Gene knockout studies have provided evidence that STAT proteins are involved in the development and function of the immune system and play a role in maintaining immune tolerance and tumor surveillance.

In the field of cell biology, TNF-related apoptosis-inducing ligand (TRAIL), is a protein functioning as a ligand that induces the process of cell death called apoptosis.

The tumor necrosis factor receptor superfamily (TNFRSF) is a protein superfamily of cytokine receptors characterized by the ability to bind tumor necrosis factors (TNFs) via an extracellular cysteine-rich domain. With the exception of nerve growth factor (NGF), all TNFs are homologous to the archetypal TNF-alpha. In their active form, the majority of TNF receptors form trimeric complexes in the plasma membrane. Accordingly, most TNF receptors contain transmembrane domains (TMDs), although some can be cleaved into soluble forms, and some lack a TMD entirely. In addition, most TNF receptors require specific adaptor protein such as TRADD, TRAF, RIP and FADD for downstream signalling. TNF receptors are primarily involved in apoptosis and inflammation, but they can also take part in other signal transduction pathways, such as proliferation, survival, and differentiation. TNF receptors are expressed in a wide variety of tissues in mammals, especially in leukocytes.
The MADS box is a conserved sequence motif. The genes which contain this motif are called the MADS-box gene family. The MADS box encodes the DNA-binding MADS domain. The MADS domain binds to DNA sequences of high similarity to the motif CC[A/T]6GG termed the CArG-box. MADS-domain proteins are generally transcription factors. The length of the MADS-box reported by various researchers varies somewhat, but typical lengths are in the range of 168 to 180 base pairs, i.e. the encoded MADS domain has a length of 56 to 60 amino acids. There is evidence that the MADS domain evolved from a sequence stretch of a type II topoisomerase in a common ancestor of all extant eukaryotes.

The tumor necrosis factor (TNF) superfamily is a protein superfamily of type II transmembrane proteins containing TNF homology domain and forming trimers. Members of this superfamily can be released from the cell membrane by extracellular proteolytic cleavage and function as a cytokine. These proteins are expressed predominantly by immune cells and they regulate diverse cell functions, including immune response and inflammation, but also proliferation, differentiation, apoptosis and embryogenesis.
Tumor necrosis factor receptor type 1-associated DEATH domain protein is a protein that in humans is encoded by the TRADD gene.

Tumor necrosis factor receptor 1 (TNFR1), also known as tumor necrosis factor receptor superfamily member 1A (TNFRSF1A) and CD120a, is a ubiquitous membrane receptor that binds tumor necrosis factor-alpha (TNFα).

Zinc finger and BTB domain-containing protein 16 is a protein that in humans is encoded by the ZBTB16 gene.

TNF receptor-associated factor 1 is a protein that in humans is encoded by the TRAF1 gene.

TNF receptor-associated factor 5 is a protein that in humans is encoded by the TRAF5 gene.

Tumor necrosis factor receptor 2 (TNFR2), also known as tumor necrosis factor receptor superfamily member 1B (TNFRSF1B) and CD120b, is one of two membrane receptors that binds tumor necrosis factor-alpha (TNFα). Like its counterpart, tumor necrosis factor receptor 1 (TNFR1), the extracellular region of TNFR2 consists of four cysteine-rich domains which allow for binding to TNFα. TNFR1 and TNFR2 possess different functions when bound to TNFα due to differences in their intracellular structures, such as TNFR2 lacking a death domain (DD).
Reticulons are a group of evolutionary conservative proteins residing predominantly in endoplasmic reticulum, primarily playing a role in promoting membrane curvature. In addition, reticulons may play a role in nuclear pore complex formation, vesicle formation, and other processes yet to be defined. They have also been linked to oligodendrocyte roles in inhibition of neurite outgrowth. Some studies link RTNs with Alzheimer's disease and amyotrophic lateral sclerosis.
Peptide signaling plays a significant role in various aspects of plant growth and development and specific receptors for various peptides have been identified as being membrane-localized receptor kinases, the largest family of receptor-like molecules in plants. Signaling peptides include members of the following protein families.

Astacins are a family of multidomain metalloendopeptidases which are either secreted or membrane-anchored. These metallopeptidases belong to the MEROPS peptidase family M12, subfamily M12A. The protein fold of the peptidase domain for members of this family resembles that of thermolysin, the type example for clan MA and the predicted active site residues for members of this family and thermolysin occur in the motif HEXXH.

Integrin-like receptors (ILRs) are found in plants and carry unique functional properties similar to true integrin proteins. True homologs of integrins exist in mammals, invertebrates, and some fungi but not in plant cells. Mammalian integrins are heterodimer transmembrane proteins that play a large role bidirectional signal transduction. As transmembrane proteins, integrins connect the extracellular matrix (ECM) to the plasma membrane of the animal cell. The extracellular matrix of plant cells, fungi, and some protist is referred to as the cell wall. The plant cell wall is composed of a tough cellulose polysaccharide rather than the collagen fibers of the animal ECM. Even with these differences, research indicates that similar proteins involved in the interaction between the ECM and animals cells are also involved in the interaction of the cell wall and plant cells.

FLS genes have been discovered to be involved in flagellin reception of bacteria. FLS1 was the original gene discovered shown to correspond with a specific ecotype within Arabidopsis thaliana. Even so, further studies have shown a second FLS gene known as FLS2 that is also associated with flagellin reception. FLS2 and FLS1 are different genes with different responsibilities, but are related genetically. FLS2 has a specific focus in plant defense and is involved in promoting the MAP kinase cascade. Mutations in the FLS2 gene can cause bacterial infection by lack of response to flg22. Therefore,FLS2’s primary focus is association with flg22 while its secondary focus is the involvement of promoting the MAP kinase cascade in plant defense.
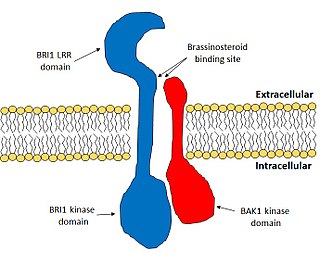
Brassinosteroid insensitive 1 (BRI1) is the major receptor of the plant hormone brassinosteroid. It plays very important roles in plant development, especially in the control of cell elongation and for the tolerance of environmental stresses. BRI1 enhances cell elongation, promotes pollen development, controls vasculature development and promotes chilling and freezing tolerance. BRI1 is one of the most well studied hormone receptors and it acts a model for the study of membrane-bound receptors in plants.

Ethylene signaling pathway is a signal transduction in plant cells to regulate important growth and developmental processes. Acting as a plant hormone, the gas ethylene is responsible for promoting the germination of seeds, ripening of fruits, the opening of flowers, the abscission of leaves and stress responses. It is the simplest alkene gas and the first gaseous molecule discovered to function as a hormone.
















