
Peripheral membrane proteins, or extrinsic membrane proteins, are membrane proteins that adhere only temporarily to the biological membrane with which they are associated. These proteins attach to integral membrane proteins, or penetrate the peripheral regions of the lipid bilayer. The regulatory protein subunits of many ion channels and transmembrane receptors, for example, may be defined as peripheral membrane proteins. In contrast to integral membrane proteins, peripheral membrane proteins tend to collect in the water-soluble component, or fraction, of all the proteins extracted during a protein purification procedure. Proteins with GPI anchors are an exception to this rule and can have purification properties similar to those of integral membrane proteins.
In biochemistry, biotinylation is the process of covalently attaching biotin to a protein, nucleic acid or other molecule. Biotinylation is rapid, specific and is unlikely to disturb the natural function of the molecule due to the small size of biotin. Biotin binds to streptavidin and avidin with an extremely high affinity, fast on-rate, and high specificity, and these interactions are exploited in many areas of biotechnology to isolate biotinylated molecules of interest. Biotin-binding to streptavidin and avidin is resistant to extremes of heat, pH and proteolysis, making capture of biotinylated molecules possible in a wide variety of environments. Also, multiple biotin molecules can be conjugated to a protein of interest, which allows binding of multiple streptavidin, avidin or neutravidin protein molecules and increases the sensitivity of detection of the protein of interest. There is a large number of biotinylation reagents available that exploit the wide range of possible labelling methods. Due to the strong affinity between biotin and streptavidin, the purification of biotinylated proteins has been a widely used approach to identify protein-protein interactions and post-translational events such as ubiquitylation in molecular biology.

The hydrophobic effect is the observed tendency of nonpolar substances to aggregate in an aqueous solution and to be excluded by water. The word hydrophobic literally means "water-fearing", and it describes the segregation of water and nonpolar substances, which maximizes the entropy of water and minimizes the area of contact between water and nonpolar molecules. In terms of thermodynamics, the hydrophobic effect is the free energy change of water surrounding a solute. A positive free energy change of the surrounding solvent indicates hydrophobicity, whereas a negative free energy change implies hydrophilicity.

Selective estrogen receptor modulators (SERMs), also known as estrogen receptor agonists/antagonists (ERAAs), are a class of drugs that act on estrogen receptors (ERs). Compared to pure ER agonists–antagonists, SERMs are more tissue-specific, allowing them to selectively inhibit or stimulate estrogen-like action in various tissues.
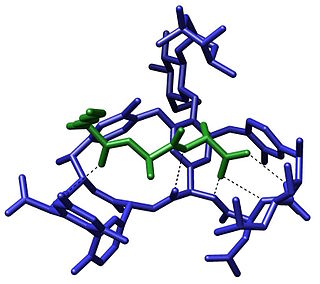
The term molecular recognition refers to the specific interaction between two or more molecules through noncovalent bonding such as hydrogen bonding, metal coordination, hydrophobic forces, van der Waals forces, π-π interactions, halogen bonding, or resonant interaction effects. In addition to these direct interactions, solvents can play a dominant indirect role in driving molecular recognition in solution. The host and guest involved in molecular recognition exhibit molecular complementarity. Exceptions are molecular containers, including, e.g., nanotubes, in which portals essentially control selectivity. Selective partioning of molecules between two or more phases can also result in molecular recognition. In partitioning-based molecular recognition the kinetics and equilibrium conditions are governed by the presence of solutes in the two phases.

Streptavidin is a 52 kDa protein (tetramer) purified from the bacterium Streptomyces avidinii. Streptavidin homo-tetramers have an extraordinarily high affinity for biotin. With a dissociation constant (Kd) on the order of ≈10−14 mol/L, the binding of biotin to streptavidin is one of the strongest non-covalent interactions known in nature. Streptavidin is used extensively in molecular biology and bionanotechnology due to the streptavidin-biotin complex's resistance to organic solvents, denaturants, detergents, proteolytic enzymes, and extremes of temperature and pH.

In biochemistry and pharmacology, a ligand is a substance that forms a complex with a biomolecule to serve a biological purpose. The etymology stems from Latin ligare, which means 'to bind'. In protein-ligand binding, the ligand is usually a molecule which produces a signal by binding to a site on a target protein. The binding typically results in a change of conformational isomerism (conformation) of the target protein. In DNA-ligand binding studies, the ligand can be a small molecule, ion, or protein which binds to the DNA double helix. The relationship between ligand and binding partner is a function of charge, hydrophobicity, and molecular structure.

In chemistry, a salt bridge is a combination of two non-covalent interactions: hydrogen bonding and ionic bonding. Ion pairing is one of the most important noncovalent forces in chemistry, in biological systems, in different materials and in many applications such as ion pair chromatography. It is a most commonly observed contribution to the stability to the entropically unfavorable folded conformation of proteins. Although non-covalent interactions are known to be relatively weak interactions, small stabilizing interactions can add up to make an important contribution to the overall stability of a conformer. Not only are salt bridges found in proteins, but they can also be found in supramolecular chemistry. The thermodynamics of each are explored through experimental procedures to access the free energy contribution of the salt bridge to the overall free energy of the state.
Cyanogen bromide is the inorganic compound with the formula (CN)Br or BrCN. It is a colorless solid that is widely used to modify biopolymers, fragment proteins and peptides, and synthesize other compounds. The compound is classified as a pseudohalogen.

In protein structures, a beta barrel(β barrel) is a beta sheet composed of tandem repeats that twists and coils to form a closed toroidal structure in which the first strand is bonded to the last strand. Beta-strands in many beta-barrels are arranged in an antiparallel fashion. Beta barrel structures are named for resemblance to the barrels used to contain liquids. Most of them are water-soluble outer membrane proteins and frequently bind hydrophobic ligands in the barrel center, as in lipocalins. Others span cell membranes and are commonly found in porins. Porin-like barrel structures are encoded by as many as 2–3% of the genes in Gram-negative bacteria. It has been shown that more than 600 proteins with various function such as oxidase, dismutase, and amylase contain the beta barrel structure.
In the fields of computational chemistry and molecular modelling, scoring functions are mathematical functions used to approximately predict the binding affinity between two molecules after they have been docked. Most commonly one of the molecules is a small organic compound such as a drug and the second is the drug's biological target such as a protein receptor. Scoring functions have also been developed to predict the strength of intermolecular interactions between two proteins or between protein and DNA.
Molecular binding is an attractive interaction between two molecules that results in a stable association in which the molecules are in close proximity to each other. It is formed when atoms or molecules bind together by sharing of electrons. It often, but not always, involves some chemical bonding.

Epiregulin (EPR) is a protein that in humans is encoded by the EREG gene.
Fragment-based lead discovery (FBLD) also known as fragment-based drug discovery (FBDD) is a method used for finding lead compounds as part of the drug discovery process. Fragments are small organic molecules which are small in size and low in molecular weight. It is based on identifying small chemical fragments, which may bind only weakly to the biological target, and then growing them or combining them to produce a lead with a higher affinity. FBLD can be compared with high-throughput screening (HTS). In HTS, libraries with up to millions of compounds, with molecular weights of around 500 Da, are screened, and nanomolar binding affinities are sought. In contrast, in the early phase of FBLD, libraries with a few thousand compounds with molecular weights of around 200 Da may be screened, and millimolar affinities can be considered useful. FBLD is a technique being used in research for discovering novel potent inhibitors. This methodology could help to design multitarget drugs for multiple diseases. The multitarget inhibitor approach is based on designing an inhibitor for the multiple targets. This type of drug design opens up new polypharmacological avenues for discovering innovative and effective therapies. Neurodegenerative diseases like Alzheimer’s (AD) and Parkinson’s, among others, also show rather complex etiopathologies. Multitarget inhibitors are more appropriate for addressing the complexity of AD and may provide new drugs for controlling the multifactorial nature of AD, stopping its progression.

Major urinary proteins (Mups), also known as α2u-globulins, are a subfamily of proteins found in abundance in the urine and other secretions of many animals. Mups provide a small range of identifying information about the donor animal, when detected by the vomeronasal organ of the receiving animal. They belong to a larger family of proteins known as lipocalins. Mups are encoded by a cluster of genes, located adjacent to each other on a single stretch of DNA, that varies greatly in number between species: from at least 21 functional genes in mice to none in humans. Mup proteins form a characteristic glove shape, encompassing a ligand-binding pocket that accommodates specific small organic chemicals.

Balanol is a fungal metabolite produced by the fungus Verticillium balanoides. It is a potent inhibitor of the serine/threonine kinases protein kinase A (PKA) and protein kinase C (PKC), binding in a similar manner with that of ATP. Balanol was discovered in 1993 in the search for novel inhibitors of PKC, a member of a family of serine/threonine kinases whose overactivation is associated with numerous human diseases of signal transduction including cancer. However, much of the research on balanol focuses on how chemical modifications of the molecular structure affect binding to PKA. Indeed, balanol, its chemically altered analogs, and their interactions with PKA in particular are used to illuminate the roles of selectivity and protein flexibility in the inhibition of kinases. For instance, the X-ray crystal structure of balanol in complex with PKA was used in order to confer selectivity and to improve pharmacological efficacy of inhibitors of the H. sapiens Akt (PKB), another serine/threonine protein kinase implicated in the proper functioning of many cellular processes.
The first antiandrogen was discovered in the 1960s. Antiandrogens antagonise the androgen receptor (AR) and thereby block the biological effects of testosterone and dihydrotestosterone (DHT). Antiandrogens are important for men with hormonally responsive diseases like prostate cancer, benign prostatic hyperplasia (BHP), acne, seborrhea, hirsutism and androgen alopecia. Antiandrogens are mainly used for the treatment of prostate diseases. Research from 2010 suggests that ARs could be linked to the disease progression of triple-negative breast cancer and salivary duct carcinoma and that antiandrogens can potentially be used to treat it.

2-sec-Butyl-4,5-dihydrothiazole (also known as SBT) is a thiazoline compound with the molecular formula C7H13NS. A volatile pheromone found in rodents such as mice and rats, SBT is excreted in the urine and promotes aggression amongst males while inducing synchronized estrus in females.
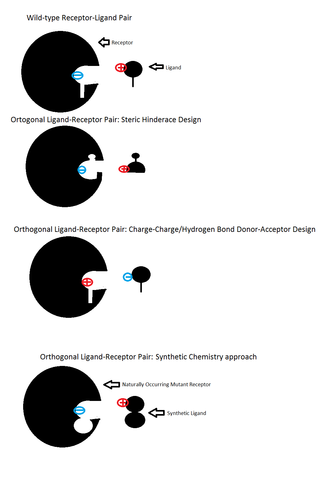
Orthogonal ligand-protein pairs are a protein-ligand binding pair made to be independent of the original binding pair. This is done by taking a mutant protein, which is activated by a different ligand. The intention here is that the orthogonal ligand will not interact with the original protein. The original protein will also be designed to not interact with the orthogonal ligand in certain cases.
The β3 adrenergic receptor agonist or β3-adrenoceptor agonist, also known as β3-AR agonist, are a class of medicine that bind selectively to β3-adrenergic receptors.













