
In chemistry, a hydrogen bond is primarily an electrostatic force of attraction between a hydrogen (H) atom which is covalently bonded to a more electronegative "donor" atom or group (Dn), and another electronegative atom bearing a lone pair of electrons—the hydrogen bond acceptor (Ac). Such an interacting system is generally denoted Dn−H···Ac, where the solid line denotes a polar covalent bond, and the dotted or dashed line indicates the hydrogen bond. The most frequent donor and acceptor atoms are the period 2 elements nitrogen (N), oxygen (O), and fluorine (F).
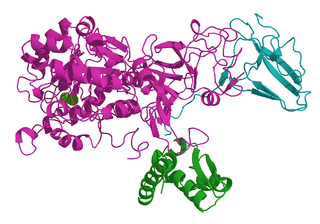
Ureases, functionally, belong to the superfamily of amidohydrolases and phosphotriesterases. Ureases are found in numerous bacteria, fungi, algae, plants, and some invertebrates, as well as in soils, as a soil enzyme. They are nickel-containing metalloenzymes of high molecular weight.
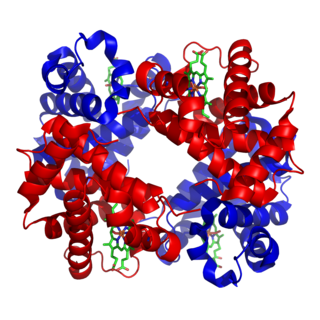
Metalloprotein is a generic term for a protein that contains a metal ion cofactor. A large proportion of all proteins are part of this category. For instance, at least 1000 human proteins contain zinc-binding protein domains although there may be up to 3000 human zinc metalloproteins.

In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate, the binding site, and residues that catalyse a reaction of that substrate, the catalytic site. Although the active site occupies only ~10–20% of the volume of an enzyme, it is the most important part as it directly catalyzes the chemical reaction. It usually consists of three to four amino acids, while other amino acids within the protein are required to maintain the tertiary structure of the enzymes.
In biochemistry, biotinylation is the process of covalently attaching biotin to a protein, nucleic acid or other molecule. Biotinylation is rapid, specific and is unlikely to disturb the natural function of the molecule due to the small size of biotin. Biotin binds to streptavidin and avidin with an extremely high affinity, fast on-rate, and high specificity, and these interactions are exploited in many areas of biotechnology to isolate biotinylated molecules of interest. Biotin-binding to streptavidin and avidin is resistant to extremes of heat, pH and proteolysis, making capture of biotinylated molecules possible in a wide variety of environments. Also, multiple biotin molecules can be conjugated to a protein of interest, which allows binding of multiple streptavidin, avidin or neutravidin protein molecules and increases the sensitivity of detection of the protein of interest. There is a large number of biotinylation reagents available that exploit the wide range of possible labelling methods. Due to the strong affinity between biotin and streptavidin, the purification of biotinylated proteins has been a widely used approach to identify protein-protein interactions and post-translational events such as ubiquitylation in molecular biology.

The hydrophobic effect is the observed tendency of nonpolar substances to aggregate in an aqueous solution and exclude water molecules. The word hydrophobic literally means "water-fearing", and it describes the segregation of water and nonpolar substances, which maximizes hydrogen bonding between molecules of water and minimizes the area of contact between water and nonpolar molecules. In terms of thermodynamics, the hydrophobic effect is the free energy change of water surrounding a solute. A positive free energy change of the surrounding solvent indicates hydrophobicity, whereas a negative free energy change implies hydrophilicity.

Lawesson's reagent (LR) is a chemical compound used in organic synthesis as a thiation agent. Lawesson's reagent was first made popular by Sven-Olov Lawesson, who did not, however, invent it. Lawesson's reagent was first made in 1956 during a systematic study of the reactions of arenes with P4S10.
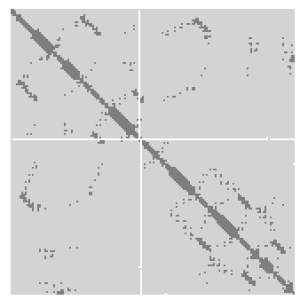
A protein contact map represents the distance between all possible amino acid residue pairs of a three-dimensional protein structure using a binary two-dimensional matrix. For two residues and , the element of the matrix is 1 if the two residues are closer than a predetermined threshold, and 0 otherwise. Various contact definitions have been proposed: The distance between the Cα-Cα atom with threshold 6-12 Å; distance between Cβ-Cβ atoms with threshold 6-12 Å ; and distance between the side-chain centers of mass.
Nitrite reductase refers to any of several classes of enzymes that catalyze the reduction of nitrite. There are two classes of NIR's. A multi haem enzyme reduces NO2− to a variety of products. Copper containing enzymes carry out a single electron transfer to produce nitric oxide.

In protein structures, a beta barrel is a beta sheet composed of tandem repeats that twists and coils to form a closed toroidal structure in which the first strand is bonded to the last strand. Beta-strands in many beta-barrels are arranged in an antiparallel fashion. Beta barrel structures are named for resemblance to the barrels used to contain liquids. Most of them are water-soluble proteins and frequently bind hydrophobic ligands in the barrel center, as in lipocalins. Others span cell membranes and are commonly found in porins. Porin-like barrel structures are encoded by as many as 2–3% of the genes in Gram-negative bacteria. It has been shown that more than 600 proteins with various function contain the beta barrel structure.

The lipocalins are a family of proteins which transport small hydrophobic molecules such as steroids, bilins, retinoids, and lipids, and most lipocalins are also able to bind to complexed iron as well as heme. They share limited regions of sequence homology and a common tertiary structure architecture. This is an eight stranded antiparallel beta barrel with a repeated + 1 topology enclosing an internal ligand binding site.
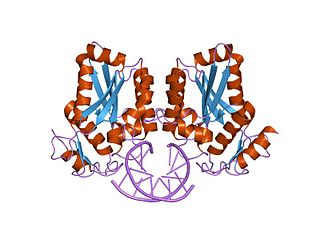
BamHI is a type II restriction endonuclease, having the capacity for recognizing short sequences of DNA and specifically cleaving them at a target site. This exhibit focuses on the structure-function relations of BamHI as described by Newman, et al. (1995). BamHI binds at the recognition sequence 5'-GGATCC-3', and cleaves these sequences just after the 5'-guanine on each strand. This cleavage results in sticky ends which are 4 bp long. In its unbound form, BamHI displays a central b sheet, which resides in between α-helices.
Carboxypeptidase A usually refers to the pancreatic exopeptidase that hydrolyzes peptide bonds of C-terminal residues with aromatic or aliphatic side-chains. Most scientists in the field now refer to this enzyme as CPA1, and to a related pancreatic carboxypeptidase as CPA2.
Cauxin is a carboxylesterase that is excreted in large amounts in cat urine. There is also evidence that it can serve as a peptide hydrolase in the production of cat pheromone precursors. Cauxin has a mass of 70 kilodaltons and is composed of 545 amino acids. The protein can also exist as a multimeric protein complex connected by disulfide bonds with a mass of 300-350 kilodaltons. This is its primary form in non-reducing conditions. The proximal tubules of epithelial cells in the kidney express cauxin. This protein is secreted into the urine from the renal tubular cells. The gene for the protein is also found in several other mammalian genomes in various organs. However, the only mammals that have cauxin present in urine are cats. It is also the first carboxylesterase to be found in urine.

A urine test strip or dipstick is a basic diagnostic tool used to determine pathological changes in a patient's urine in standard urinalysis.

Major urinary proteins (Mups), also known as α2u-globulins, are a subfamily of proteins found in abundance in the urine and other secretions of many animals. Mups provide a small range of identifying information about the donor animal, when detected by the vomeronasal organ of the receiving animal. They belong to a larger family of proteins known as lipocalins. Mups are encoded by a cluster of genes, located adjacent to each other on a single stretch of DNA, that varies greatly in number between species: from at least 21 functional genes in mice to none in humans. Mup proteins form a characteristic glove shape, encompassing a ligand-binding pocket that accommodates specific small organic chemicals.

Balanol is a fungal metabolite produced by the fungus Verticillium balanoides. It is a potent inhibitor of the serine/threonine kinases protein kinase A (PKA) and protein kinase C (PKC), binding in a similar manner with that of ATP. Balanol was discovered in 1993 in the search for novel inhibitors of PKC, a member of a family of serine/threonine kinases whose overactivation is associated with numerous human diseases of signal transduction including cancer. However, much of the research on balanol focuses on how chemical modifications of the molecular structure affect binding to PKA. Indeed, balanol, its chemically altered analogs, and their interactions with PKA in particular are used to illuminate the roles of selectivity and protein flexibility in the inhibition of kinases. For instance, the X-ray crystal structure of balanol in complex with PKA was used in order to confer selectivity and to improve pharmacological efficacy of inhibitors of the H. sapiens Akt (PKB), another serine/threonine protein kinase implicated in the proper functioning of many cellular processes.

N-Phenylnaphthalen-1-amine (NPN) is an aromatic amine with the chemical formula C
16H
12NH.
Many bacteria secrete small iron-binding molecules called siderophores, which bind strongly to ferric ions. FepA is an integral bacterial outer membrane porin protein that belongs to outer membrane receptor family and provides the active transport of iron bound by the siderophore enterobactin from the extracellular space, into the periplasm of Gram-negative bacteria. FepA has also been shown to transport vitamin B12, and colicins B and D as well. This protein belongs to family of ligand-gated protein channels.
Mercury(II) reductase (EC 1.16.1.1), commonly known as MerA, is an oxidoreductase enzyme and flavoprotein that catalyzes the reduction of Hg2+ to Hg0. Mercury(II) reductase is found in the cytoplasm of many eubacteria in both aerobic and anaerobic environments and serves to convert toxic mercury ions into relatively inert elemental mercury.
















