| |
| Identifiers | |
|---|---|
3D model (JSmol) | |
| ChEBI | |
| ChEMBL | |
| ChemSpider | |
PubChem CID | |
| |
| |
| Properties | |
| C40H46ClNO9 | |
| Molar mass | 720.26 g·mol−1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
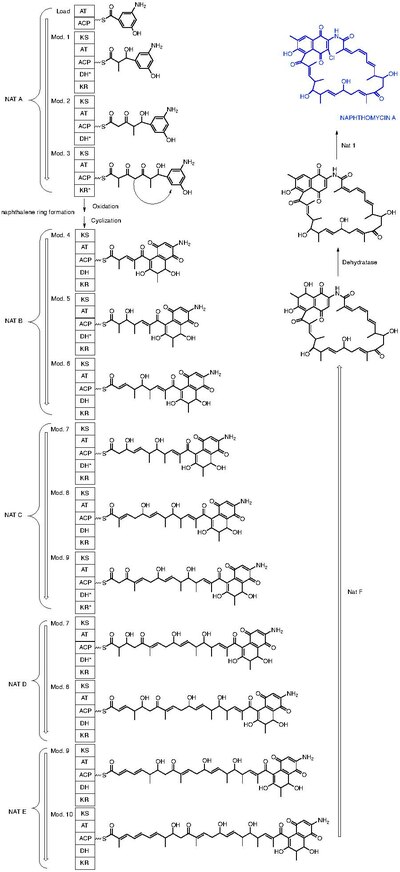
Naphthomycin A is a type of naphthomycin. It was isolated as a yellow pigment from Streptomyces collinus and it shows antibacterial, antifungal, and antitumor activities. [1] Naphthomycins have the longest ansa aliphatic carbon chain of the ansamycin family. [2] Biosynthetic origins of the carbon skeleton from PKS1 were investigated by feeding 13C-labeled precursors and subsequent 13C-NMR product analysis. Naphthomycin gene clusters have been cloned and sequenced to confirm involvement in biosynthesis via deletion of a 7.2kb region. [3] Thirty-two genes were identified in the 106kb cluster. [4]