Related Research Articles
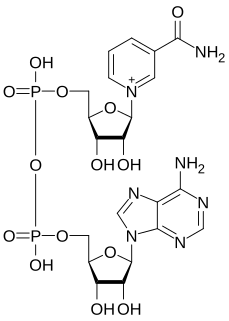
Nicotinamide adenine dinucleotide (NAD) is a coenzyme central to metabolism. Found in all living cells, NAD is called a dinucleotide because it consists of two nucleotides joined through their phosphate groups. One nucleotide contains an adenine nucleobase and the other nicotinamide. NAD exists in two forms: an oxidized and reduced form, abbreviated as NAD+ and NADH (H for hydrogen), respectively.

The bacteria capsule is a large structure common to many bacteria. It is a polysaccharide layer that lies outside the cell envelope, and is thus deemed part of the outer envelope of a bacterial cell. It is a well-organized layer, not easily washed off, and it can be the cause of various diseases.
Magnesium transporters are proteins that transport magnesium across the cell membrane. All forms of life require magnesium, yet the molecular mechanisms of Mg2+ uptake from the environment and the distribution of this vital element within the organism are only slowly being elucidated.
Sirtuins are a family of signaling proteins involved in metabolic regulation. They are ancient in animal evolution and appear to possess a highly conserved structure throughout all kingdoms of life. Chemically, sirtuins are a class of proteins that possess either mono-ADP-ribosyltransferase or deacylase activity, including deacetylase, desuccinylase, demalonylase, demyristoylase and depalmitoylase activity. The name Sir2 comes from the yeast gene 'silent mating-type information regulation 2', the gene responsible for cellular regulation in yeast.

The gcvB RNA gene encodes a small non-coding RNA involved in the regulation of a number of amino acid transport systems as well as amino acid biosynthetic genes. The GcvB gene is found in enteric bacteria such as Escherichia coli. GcvB regulates genes by acting as an antisense binding partner of the mRNAs for each regulated gene. This binding is dependent on binding to a protein called Hfq. Transcription of the GcvB RNA is activated by the adjacent GcvA gene and repressed by the GcvR gene. A deletion of GcvB RNA from Y. pestis changed colony shape as well as reducing growth. It has been shown by gene deletion that GcvB is a regulator of acid resistance in E. coli. GcvB enhances the ability of the bacterium to survive low pH by upregulating the levels of the alternate sigma factor RpoS. A polymeric form of GcvB has recently been identified. Interaction of GcvB with small RNA SroC triggers the degradation of GcvB by RNase E, lifting the GcvB-mediated mRNA repression of its target genes.

NAD+ kinase (EC 2.7.1.23, NADK) is an enzyme that converts nicotinamide adenine dinucleotide (NAD+) into NADP+ through phosphorylating the NAD+ coenzyme. NADP+ is an essential coenzyme that is reduced to NADPH primarily by the pentose phosphate pathway to provide reducing power in biosynthetic processes such as fatty acid biosynthesis and nucleotide synthesis. The structure of the NADK from the archaean Archaeoglobus fulgidus has been determined.
In enzymology, a D-malate dehydrogenase (decarboxylating) (EC 1.1.1.83) is an enzyme that catalyzes the chemical reaction
In enzymology, a histidinol dehydrogenase (HIS4) (HDH) (EC 1.1.1.23) is an enzyme that catalyzes the chemical reaction

Nicotinamide phosphoribosyltransferase, formerly known as pre-B-cell colony-enhancing factor 1 (PBEF1) or visfatin for its extracellular form (eNAMPT), is an enzyme that in humans is encoded by the NAMPT gene. The intracellular form of this protein (iNAMPT) is the rate-limiting enzyme in the nicotinamide adenine dinucleotide (NAD+) salvage pathway that converts nicotinamide to nicotinamide mononucleotide (NMN) which is responsible for most of the NAD+ formation in mammals. iNAMPT can also catalyze the synthesis of NMN from phosphoribosyl pyrophosphate (PRPP) when ATP is present. eNAMPT has been reported to be a cytokine (PBEF) that activates TLR4, that promotes B cell maturation, and that inhibits neutrophil apoptosis.

In enzymology, a hydroxyethylthiazole kinase is an enzyme that catalyzes the chemical reaction

Bacterial microcompartments (BMCs) are organelle-like structures found in bacteria. They consist of a protein shell that encloses enzymes and other proteins. BMCs are typically about 40–200 nanometers in diameter and are made entirely of proteins. The shell functions like a membrane, as it is selectively permeable. Other protein-based compartments found in bacteria and archaea include encapsulin nanocompartments and gas vesicles.

Autoinducer-2 (AI-2), a furanosyl borate diester or tetrahydroxy furan, is a member of a family of signaling molecules used in quorum sensing. AI-2 is one of only a few known biomolecules incorporating boron. First identified in the marine bacterium Vibrio harveyi, AI-2 is produced and recognized by many Gram-negative and Gram-positive bacteria. AI-2 arises by the reaction of 4,5-Dihydroxy-2,3-pentanedione, which is produced enzymatically with boric acid, and is recognized by the two-component sensor kinase LuxPQ in Vibrionaceae.
Tripartite ATP-independent periplasmic transporters are a large family of solute transporters found in bacteria and archaea, but not in eukaryotes, that appear to be specific for the uptake of organic acids or related molecules containing a carboxylate or sulfonate group. They are unique in that they utilize a substrate binding protein (SBP) in combination with a secondary transporter.
The alpha-D-phosphohexomutases are a large superfamily of enzymes, with members in all three domains of life. Enzymes from this superfamily are ubiquitous in organisms from E. Coli to humans, and catalyze a phosphoryl transfer reaction on a phosphosugar substrate. Four well studied subgroups in the superfamily are:
- Phosphoglucomutase (PGM)
- Phosphoglucomutase/Phosphomannomutase (PGM/PMM)
- Phosphoglucosamine mutase (PNGM)
- Phosphoaceytlglucosamine mutase (PAGM)
D-glycero-beta-D-manno-heptose-7-phosphate kinase is an enzyme with systematic name ATP:D-glycero-beta-D-manno-heptose 7-phosphate 1-phosphotransferase. This enzyme catalyses the following chemical reaction

Nicotinamide riboside (NR, SR647) is a pyridine-nucleoside similar to vitamin B3, functioning as a precursor to nicotinamide adenine dinucleotide or NAD+.
The multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) flippase superfamily is a group of integral membrane protein families. The MOP flippase superfamily includes twelve distantly related families, six for which functional data are available:
- One ubiquitous family (MATE) specific for drugs - (TC# 2.A.66.1) The Multi Antimicrobial Extrusion (MATE) Family
- One (PST) specific for polysaccharides and/or their lipid-linked precursors in prokaryotes - (TC# 2.A.66.2) The Polysaccharide Transport (PST) Family
- One (OLF) specific for lipid-linked oligosaccharide precursors of glycoproteins in eukaryotes - (TC# 2.A.66.3) The Oligosaccharidyl-lipid Flippase (OLF) Family
- One (MVF) lipid-peptidoglycan precursor flippase involved in cell wall biosynthesis - (TC# 2.A.66.4) The Mouse Virulence Factor (MVF) Family
- One (AgnG) which includes a single functionally characterized member that extrudes the antibiotic, Agrocin 84 - (TC# 2.A.66.5) The Agrocin 84 Antibiotic Exporter (AgnG) Family
- And finally, one (Ank) that shuttles inorganic pyrophosphate (PPi) - (TC# 2.A.66.9) The Progressive Ankylosis (Ank) Family
The C4-dicarboxylate uptake family or Dcu family is a family of transmembrane ion transporters found in bacteria. Their function is to exchange dicarboxylates such as aspartate, malate, fumarate and succinate.

Nicotinamide mononucleotide is a nucleotide derived from ribose, nicotinamide, nicotinamide riboside and niacin. Humans have enzymes that can use NMN to generate nicotinamide adenine dinucleotide (NADH). In mice, NMN has been proposed to enter cells via the small intestines within 10 minutes converting to NAD+ through the Slc12a8 transporter. However this observation has been challenged, and remains unsettled.
N-glycosyltransferase is an enzyme in prokaryotes which transfers individual hexoses onto asparagine sidechains in substrate proteins, using a nucleotide-bound intermediary, within the cytoplasm. They are distinct from regular N-glycosylating enzymes, which are oligosaccharyltransferases that transfer pre-assembled oligosaccharides. Both enzyme families however target a shared amino acid sequence asparagine—-any amino acid except proline—serine or threonine (N–x–S/T), with some variations.
References
- 1 2 Yee DC, Shlykov MA, Västermark A, Reddy VS, Arora S, Sun EI, Saier MH (November 2013). "The transporter-opsin-G protein-coupled receptor (TOG) superfamily". The FEBS Journal. 280 (22): 5780–800. doi:10.1111/febs.12499. PMC 3832197 . PMID 23981446.
- 1 2 Saier, MH Jr. "4.B.1 The Nicotinamide Ribonucleoside (NR) Uptake Permease (PnuC) Family". Transporter Classification Database. Saier Lab Bioinformatics Group / SDSC.
- ↑ Foster JW, Park YK, Penfound T, Fenger T, Spector MP (August 1990). "Regulation of NAD metabolism in Salmonella typhimurium: molecular sequence analysis of the bifunctional nadR regulator and the nadA-pnuC operon". Journal of Bacteriology. 172 (8): 4187–96. doi:10.1128/jb.172.8.4187-4196.1990. PMC 213241 . PMID 2198247.
- 1 2 Merdanovic M, Sauer E, Reidl J (July 2005). "Coupling of NAD+ biosynthesis and nicotinamide ribosyl transport: characterization of NadR ribonucleotide kinase mutants of Haemophilus influenzae". Journal of Bacteriology. 187 (13): 4410–20. doi:10.1128/JB.187.13.4410-4420.2005. PMC 1151767 . PMID 15968050.
- ↑ Penfound T, Foster JW (January 1999). "NAD-dependent DNA-binding activity of the bifunctional NadR regulator of Salmonella typhimurium". Journal of Bacteriology. 181 (2): 648–55. doi:10.1128/JB.181.2.648-655.1999. PMC 93422 . PMID 9882682.
- ↑ Mitchell P, Moyle J (August 1958). "Group-translocation: a consequence of enzyme-catalysed group-transfer". Nature. 182 (4632): 372–3. Bibcode:1958Natur.182..372M. doi:10.1038/182372a0. PMID 13577842. S2CID 4261342.
- 1 2 Kurnasov OV, Polanuyer BM, Ananta S, Sloutsky R, Tam A, Gerdes SY, Osterman AL (December 2002). "Ribosylnicotinamide kinase domain of NadR protein: identification and implications in NAD biosynthesis". Journal of Bacteriology. 184 (24): 6906–17. doi:10.1128/jb.184.24.6906-6917.2002. PMC 135457 . PMID 12446641.
- ↑ Cynamon MH, Sorg TB, Patapow A (October 1988). "Utilization and metabolism of NAD by Haemophilus parainfluenzae". Journal of General Microbiology. 134 (10): 2789–99. doi: 10.1099/00221287-134-10-2789 . PMID 3254936.
- ↑ Kemmer G, Reilly TJ, Schmidt-Brauns J, Zlotnik GW, Green BA, Fiske MJ, Herbert M, Kraiss A, Schlör S, Smith A, Reidl J (July 2001). "NadN and e (P4) are essential for utilization of NAD and nicotinamide mononucleotide but not nicotinamide riboside in Haemophilus influenzae". Journal of Bacteriology. 183 (13): 3974–81. doi:10.1128/JB.183.13.3974-3981.2001. PMC 95280 . PMID 11395461.
- ↑ Reidl J, Schlör S, Kraiss A, Schmidt-Brauns J, Kemmer G, Soleva E (March 2000). "NADP and NAD utilization in Haemophilus influenzae". Molecular Microbiology. 35 (6): 1573–81. doi:10.1046/j.1365-2958.2000.01829.x. PMID 10760156. S2CID 29776509.
- ↑ Schmidt-Brauns J, Herbert M, Kemmer G, Kraiss A, Schlör S, Reidl J (August 2001). "Is a NAD pyrophosphatase activity necessary for Haemophilus influenzae type b multiplication in the blood stream?". International Journal of Medical Microbiology. 291 (3): 219–25. doi:10.1078/1438-4221-00122. PMID 11554562.
- ↑ Andersen C, Maier E, Kemmer G, Blass J, Hilpert AK, Benz R, Reidl J (July 2003). "Porin OmpP2 of Haemophilus influenzae shows specificity for nicotinamide-derived nucleotide substrates". The Journal of Biological Chemistry. 278 (27): 24269–76. doi: 10.1074/jbc.M213087200 . PMID 12695515.
- ↑ Herbert M, Sauer E, Smethurst G, Kraiss A, Hilpert AK, Reidl J (September 2003). "Nicotinamide ribosyl uptake mutants in Haemophilus influenzae". Infection and Immunity. 71 (9): 5398–401. doi:10.1128/iai.71.9.5398-5401.2003. PMC 187334 . PMID 12933892.
- ↑ Sauer E, Merdanovic M, Mortimer AP, Bringmann G, Reidl J (December 2004). "PnuC and the utilization of the nicotinamide riboside analog 3-aminopyridine in Haemophilus influenzae". Antimicrobial Agents and Chemotherapy. 48 (12): 4532–41. doi:10.1128/AAC.48.12.4532-4541.2004. PMC 529221 . PMID 15561822.
- ↑ Singh SK, Kurnasov OV, Chen B, Robinson H, Grishin NV, Osterman AL, Zhang H (September 2002). "Crystal structure of Haemophilus influenzae NadR protein. A bifunctional enzyme endowed with NMN adenyltransferase and ribosylnicotinimide kinase activities". The Journal of Biological Chemistry. 277 (36): 33291–9. doi: 10.1074/jbc.M204368200 . PMID 12068016.
- 1 2 Foster JW, Penfound T (September 1993). "The bifunctional NadR regulator of Salmonella typhimurium: location of regions involved with DNA binding, nucleotide transport and intramolecular communication". FEMS Microbiology Letters. 112 (2): 179–83. doi: 10.1016/0378-1097(93)90161-t . PMID 8405960.
- ↑ Tempel W, Rabeh WM, Bogan KL, Belenky P, Wojcik M, Seidle HF, Nedyalkova L, Yang T, Sauve AA, Park HW, Brenner C (October 2007). "Nicotinamide riboside kinase structures reveal new pathways to NAD+". PLOS Biology. 5 (10): e263. doi:10.1371/journal.pbio.0050263. PMC 1994991 . PMID 17914902.
- ↑ Jaehme M, Guskov A, Slotboom DJ (February 2016). "Pnu Transporters: Ain't They SWEET?" (PDF). Trends in Biochemical Sciences. 41 (2): 117–118. doi:10.1016/j.tibs.2015.11.013. PMID 26692123.