
A protein kinase is a kinase which selectively modifies other proteins by covalently adding phosphates to them (phosphorylation) as opposed to kinases which modify lipids, carbohydrates, or other molecules. Phosphorylation usually results in a functional change of the target protein (substrate) by changing enzyme activity, cellular location, or association with other proteins. The human genome contains about 500 protein kinase genes and they constitute about 2% of all human genes. There are two main types of protein kinase. The great majority are serine/threonine kinases, which phosphorylate the hydroxyl groups of serines and threonines in their targets. Most of the others are tyrosine kinases, although additional types exist. Protein kinases are also found in bacteria and plants. Up to 30% of all human proteins may be modified by kinase activity, and kinases are known to regulate the majority of cellular pathways, especially those involved in signal transduction.

Aspartate carbamoyltransferase catalyzes the first step in the pyrimidine biosynthetic pathway.
An NTP binding site is a type of binding site found in nucleoside monophosphate (NMP) kinases, N can be adenosine or guanosine. A P-loop is one of the structural motifs common for nucleoside triphosphate (NTP) binding sites, it interacts with the bound nucleotide's phosphoryl groups. For the binding site to be able to bind a nucleotide, the nucleotide must be complex bound to Mg2+ or Mn2+. Nucleotide binding will cause conformational changes in the protein because the P-loop will bend.
Biosynthesis, i.e., chemical synthesis occurring in biological contexts, is a term most often referring to multi-step, enzyme-catalyzed processes where chemical substances absorbed as nutrients serve as enzyme substrates, with conversion by the living organism either into simpler or more complex products. Examples of biosynthetic pathways include those for the production of amino acids, lipid membrane components, and nucleotides, but also for the production of all classes of biological macromolecules, and of acetyl-coenzyme A, adenosine triphosphate, nicotinamide adenine dinucleotide and other key intermediate and transactional molecules needed for metabolism. Thus, in biosynthesis, any of an array of compounds, from simple to complex, are converted into other compounds, and so it includes both the catabolism and anabolism of complex molecules. Biosynthetic processes are often represented via charts of metabolic pathways. A particular biosynthetic pathway may be located within a single cellular organelle, while others involve enzymes that are located across an array of cellular organelles and structures.
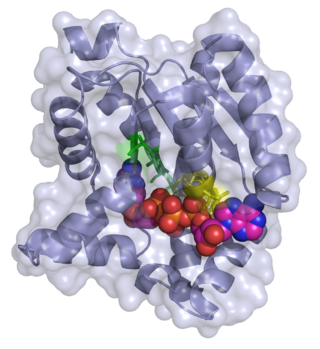
Adenylate kinase is a phosphotransferase enzyme that catalyzes the interconversion of the various adenosine phosphates. By constantly monitoring phosphate nucleotide levels inside the cell, ADK plays an important role in cellular energy homeostasis.
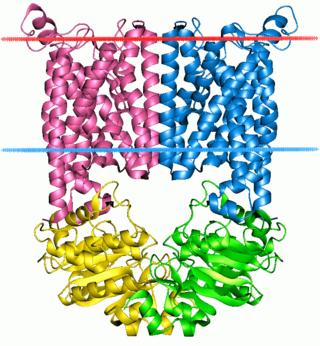
The ABC transporters, ATP synthase (ATP)-binding cassette transporters are a transport system superfamily that is one of the largest and possibly one of the oldest gene families. It is represented in all extant phyla, from prokaryotes to humans. ABC transporters belong to translocases.

An ATP-binding motif is a 250-residue sequence within an ATP-binding protein’s primary structure. The binding motif is associated with a protein’s structure and/or function. ATP is a molecule of energy, and can be a coenzyme, involved in a number of biological reactions. ATP is proficient at interacting with other molecules through a binding site. The ATP binding site is the environment in which ATP catalytically actives the enzyme and, as a result, is hydrolyzed to ADP. The binding of ATP causes a conformational change to the enzyme it is interacting with.

Amino acid biosynthesis is the set of biochemical processes by which the amino acids are produced. The substrates for these processes are various compounds in the organism's diet or growth media. Not all organisms are able to synthesize all amino acids. For example, humans can synthesize 11 of the 20 standard amino acids. These 11 are called the non-essential amino acids.
Pantothenate kinase (EC 2.7.1.33, PanK; CoaA) is the first enzyme in the Coenzyme A (CoA) biosynthetic pathway. It phosphorylates pantothenate (vitamin B5) to form 4'-phosphopantothenate at the expense of a molecule of adenosine triphosphate (ATP). It is the rate-limiting step in the biosynthesis of CoA.

BRAF is a human gene that encodes a protein called B-Raf. The gene is also referred to as proto-oncogene B-Raf and v-Raf murine sarcoma viral oncogene homolog B, while the protein is more formally known as serine/threonine-protein kinase B-Raf.

Diphosphomevalonate decarboxylase (EC 4.1.1.33), most commonly referred to in scientific literature as mevalonate diphosphate decarboxylase, is an enzyme that catalyzes the chemical reaction
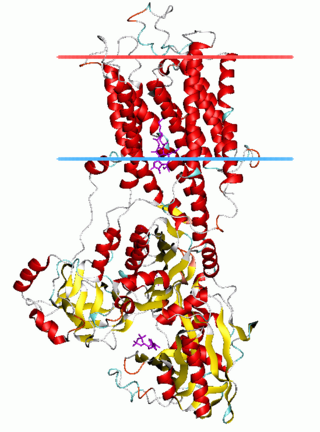
The P-type ATPases, also known as E1-E2 ATPases, are a large group of evolutionarily related ion and lipid pumps that are found in bacteria, archaea, and eukaryotes. P-type ATPases are α-helical bundle primary transporters named based upon their ability to catalyze auto- (or self-) phosphorylation (hence P) of a key conserved aspartate residue within the pump and their energy source, adenosine triphosphate (ATP). In addition, they all appear to interconvert between at least two different conformations, denoted by E1 and E2. P-type ATPases fall under the P-type ATPase (P-ATPase) Superfamily (TC# 3.A.3) which, as of early 2016, includes 20 different protein families.

Histidine kinases (HK) are multifunctional, and in non-animal kingdoms, typically transmembrane, proteins of the transferase class of enzymes that play a role in signal transduction across the cellular membrane. The vast majority of HKs are homodimers that exhibit autokinase, phosphotransfer, and phosphatase activity. HKs can act as cellular receptors for signaling molecules in a way analogous to tyrosine kinase receptors (RTK). Multifunctional receptor molecules such as HKs and RTKs typically have portions on the outside of the cell that bind to hormone- or growth factor-like molecules, portions that span the cell membrane, and portions within the cell that contain the enzymatic activity. In addition to kinase activity, the intracellular domains typically have regions that bind to a secondary effector molecule or complex of molecules that further propagate signal transduction within the cell. Distinct from other classes of protein kinases, HKs are usually parts of a two-component signal transduction mechanisms in which HK transfers a phosphate group from ATP to a histidine residue within the kinase, and then to an aspartate residue on the receiver domain of a response regulator protein. More recently, the widespread existence of protein histidine phosphorylation distinct from that of two-component histidine kinases has been recognised in human cells. In marked contrast to Ser, Thr and Tyr phosphorylation, the analysis of phosphorylated Histidine using standard biochemical and mass spectrometric approaches is much more challenging, and special procedures and separation techniques are required for their preservation alongside classical Ser, Thr and Tyr phosphorylation on proteins isolated from human cells.
In enzymology, a nucleoside-phosphate kinase is an enzyme that catalyzes the chemical reaction
Eukaryotic Initiation Factor 2 (eIF2) is an eukaryotic initiation factor. It is required for most forms of eukaryotic translation initiation. eIF2 mediates the binding of tRNAiMet to the ribosome in a GTP-dependent manner. eIF2 is a heterotrimer consisting of an alpha, a beta, and a gamma subunit.

Pho4 is a protein with a basic helix-loop-helix (bHLH) transcription factor. It is found in S. cerevisiae and other yeasts. It functions as a transcription factor to regulate phosphate responsive genes located in yeast cells. The Pho4 protein homodimer is able to do this by binding to DNA sequences containing the bHLH binding site 5'-CACGTG-3'. This sequence is found in the promoters of genes up-regulated in response to phosphate availability such as the PHO5 gene.

The WRKY domain is found in the WRKY transcription factor family, a class of transcription factors. The WRKY domain is found almost exclusively in plants although WRKY genes appear present in some diplomonads, social amoebae and other amoebozoa, and fungi incertae sedis. They appear absent in other non-plant species. WRKY transcription factors have been a significant area of plant research for the past 20 years. The WRKY DNA-binding domain recognizes the W-box (T)TGAC(C/T) cis-regulatory element.
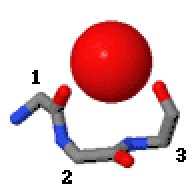
In the area of protein structural motifs, niches are three or four amino acid residue features in which main-chain CO groups are bridged by positively charged or δ+ groups. The δ+ groups include groups with two hydrogen bond donor atoms such as NH2 groups and water molecules. In typical proteins, 7% of amino acid residues belong to niches bound to a δ+ group, while another 7% have the conformation but no single cationic bridging group is detected.
Eps15 homology domain-containing protein 3, abbreviated as EHD3 and also known as PAST3, is a protein encoded by the EHD3 gene. It has been observed in humans, mice and rats. It belongs to the EHD protein family, a group of four membrane remodeling proteins related to the Dynamin superfamily of large GTPases. Although the four of them are 70-80% amino acid identical, they all have different locations. Its main function is related to endocytic transport.
In molecular biology, an arginine finger is an amino acid residue of some enzymes. Arginine fingers are often found in the protein superfamily of AAA+ ATPases, GTPases, and dUTPases, where they assist in the catalysis of the gamma phosphate or gamma and beta phosphates from ATP or GTP, which creates a release of energy which can be used to perform cellular work. They are also found in GTPase-activating proteins (GAP). Thus, they are essential for many forms of life, and are highly conserved. Arginine fingers function through non-covalent interactions. They may also assist in dimerization, and while they are found in a wide variety of enzymes, they are not ubiquitous.













