
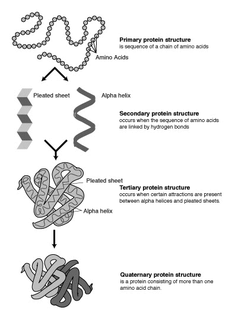
The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand-helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located three or four residues earlier along the protein sequence.

The β-sheet is a common motif of regular secondary structure in proteins. Beta sheets consist of beta strands connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet. A β-strand is a stretch of polypeptide chain typically 3 to 10 amino acids long with backbone in an extended conformation. The supramolecular association of β-sheets has been implicated in formation of the protein aggregates and fibrils observed in many human diseases, notably the amyloidoses such as Alzheimer's disease.

In biology, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate and residues that catalyse a reaction of that substrate. Although the active site occupies only ~10–20% of the volume of an enzyme, it is the most important part as it directly catalyzes the chemical reaction. It usually consists of three to four amino acids, while other amino acids within the protein are required to maintain the tertiary structure of the enzyme.
Protein structure prediction is the inference of the three-dimensional structure of a protein from its amino acid sequence—that is, the prediction of its folding and its secondary and tertiary structure from its primary structure. Structure prediction is fundamentally different from the inverse problem of protein design. Protein structure prediction is one of the most important goals pursued by bioinformatics and theoretical chemistry; it is highly important in medicine and biotechnology. Every two years, the performance of current methods is assessed in the CASP experiment. A continuous evaluation of protein structure prediction web servers is performed by the community project CAMEO3D.
In a chain-like biological molecule, such as a protein or nucleic acid, a structural motif is a supersecondary structure, which also appears in a variety of other molecules. Motifs do not allow us to predict the biological functions: they are found in proteins and enzymes with dissimilar functions.

An ATP, adenosine triphosphate, binding motif is a 250 residue sequence within an ATP binding protein’s primary structure. The binding motif is associated with a protein’s structure and/or function. ATP is a molecule of energy, and can be a coenzyme, involved in a number of biological reactions. ATP is proficient at interacting with other molecules through a binding site. The ATP binding site is the environment in which ATP catalytically actives the enzyme and, as a result, is hydrolyzed to ADP. The binding of ATP causes a conformational change to the enzyme it is interacting with.
A turn is an element of secondary structure in proteins where the polypeptide chain reverses its overall direction.
A beta bulge can be described as a localized disruption of the regular hydrogen bonding of beta sheet by inserting extra residues into one or both hydrogen bonded β-strands.

The beta hairpin is a simple protein structural motif involving two beta strands that look like a hairpin. The motif consists of two strands that are adjacent in primary structure, oriented in an antiparallel direction, and linked by a short loop of two to five amino acids. Beta hairpins can occur in isolation or as part of a series of hydrogen bonded strands that collectively comprise a beta sheet.

The EF hand is a helix-loop-helix structural domain or motif found in a large family of calcium-binding proteins.
β turns are the most common form of turns—a type of non-regular secondary structure in proteins that cause a change in direction of the polypeptide chain. They are very common motifs in proteins and polypeptides. Each consists of four amino acid residues. They can be defined in two ways: 1. By the possession of an intra-main-chain hydrogen bond between the CO of residue i and the NH of residue i+3; Alternatively, 2. By having a distance of less than 7Å between the Cα atoms of residues i and i+3. The hydrogen bond criterion is the one most appropriate for everyday use, partly because it gives rise to four distinct categories; the distance criterion gives rise to the same four categories but yields additional turn types.
The Walker A and Walker B motifs are protein sequence motifs, known to have highly conserved three-dimensional structures. These were first reported in ATP-binding proteins by Walker and co-workers in 1982.
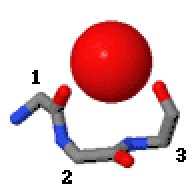
In the area of protein structural motifs, niches are three or four amino acid residue features in which main-chain CO groups are bridged by positively charged or δ+ groups. The δ+ groups include groups with two hydrogen bond donor atoms such as NH2 groups and water molecules. In typical proteins, 7% of amino acid residues belong to niches bound to a δ+ group, while another 7% have the conformation but no single cationic bridging group is detected.

Schellman loops are commonly occurring structural features of proteins and polypeptides. Each has six amino acid residues with two specific inter-mainchain hydrogen bonds and a characteristic main chain dihedral angle conformation. The CO group of residue i is hydrogen-bonded to the NH of residue i+5, and the CO group of residue i+1 is hydrogen-bonded to the NH of residue i+4. Residues i+1, i+2, and i+3 have negative φ (phi) angle values and the phi value of residue i+4 is positive. Schellman loops incorporate a three amino acid residue RL nest, in which three mainchain NH groups form a concavity for hydrogen bonding to carbonyl oxygens. About 2.5% of amino acids in proteins belong to Schellman loops. Two websites are available for examining small motifs in proteins, Motivated Proteins: ; or PDBeMotif:.
The Asx turn is a structural feature in proteins and polypeptides. It consists of three amino acid residues in which residue i is an aspartate (Asp) or asparagine (Asn) that forms a hydrogen bond from its sidechain CO group to the mainchain NH group of residue i+2. About 14% of Asx residues present in proteins belong to Asx turns.

Catgrips are small cation-binding molecular features of proteins and peptides. Each consists of the main chain atoms only of three consecutive amino acid residues. The first and third main chain CO groups bind the cations, often calcium, magnesium, potassium or sodium, with no side chain involvement. Many catgrips bind a water molecule instead of a cation; it is hydrogen-bonded to the first and third main chain CO groups. Catgrips are found as calcium-binding features in annexins, matrix metalloproteinases (e.g.serralysins), subtilisins and phospholipase A2. They are also observed in synthetic peptides and in cyclic hexapeptides made from alternating D,L amino acids.

The Asx motif is a commonly occurring feature in proteins and polypeptides. It consists of four or five amino acid residues with either aspartate or asparagine as the first residue. It is defined by two internal hydrogen bonds. One is between the side chain oxygen of residue i and the main chain NH of residue i+2 or i+3; the other is between the main chain oxygen of residue i and the main chain NH of residue i+3 or i+4. Asx motifs occur commonly in proteins and polypeptides.

The ST motif is a commonly occurring feature in proteins and polypeptides. It consists of four or five amino acid residues with either serine or threonine as the first residue. It is defined by two internal hydrogen bonds. One is between the side chain oxygen of residue i and the main chain NH of residue i + 2 or i + 3; the other is between the main chain oxygen of residue i and the main chain NH of residue i + 3 or i + 4. Two websites are available for finding and examining ST motifs in proteins, Motivated Proteins: and PDBeMotif.
Amide Rings are small motifs in proteins and polypeptides. They consist of 9-atom or 11-atom rings formed by two CO...HN hydrogen bonds between a side chain amide group and the main chain atoms of a short polypeptide. They are observed with glutamine or asparagine side chains within proteins and polypeptides. Structurally similar rings occur in the binding of purine, pyrimidine and nicotinamide bases to the main chain atoms of proteins. About 4% of asparagines and glutamines form amide rings; in databases of protein domain structures, one is present, on average, every other protein.

Beta bulge loops are commonly occurring motifs in proteins and polypeptides consisting of five to six amino acids. There are two types: type 1, which is a pentapeptide; and type 2, with six amino acids. They are regarded as a type of beta bulge, and have the alternative name of type G1 beta bulge. Compared to other beta bulges, beta bulge loops give rise to chain reversal such that they often occur at the loop ends of beta hairpins; hairpins of this sort can be described as 3:5 or 4:6. Two websites are available for finding and examining β bulge loops in proteins, Motivated Proteins: and PDBeMotif:.













