Cell biology is a branch of biology that studies the structure, function, and behavior of cells. All living organisms are made of cells. A cell is the basic unit of life that is responsible for the living and functioning of organisms. Cell biology is the study of the structural and functional units of cells. Cell biology encompasses both prokaryotic and eukaryotic cells and has many subtopics which may include the study of cell metabolism, cell communication, cell cycle, biochemistry, and cell composition. The study of cells is performed using several microscopy techniques, cell culture, and cell fractionation. These have allowed for and are currently being used for discoveries and research pertaining to how cells function, ultimately giving insight into understanding larger organisms. Knowing the components of cells and how cells work is fundamental to all biological sciences while also being essential for research in biomedical fields such as cancer, and other diseases. Research in cell biology is interconnected to other fields such as genetics, molecular genetics, molecular biology, medical microbiology, immunology, and cytochemistry.

Autophagy is the natural, conserved degradation of the cell that removes unnecessary or dysfunctional components through a lysosome-dependent regulated mechanism. It allows the orderly degradation and recycling of cellular components. Although initially characterized as a primordial degradation pathway induced to protect against starvation, it has become increasingly clear that autophagy also plays a major role in the homeostasis of non-starved cells. Defects in autophagy have been linked to various human diseases, including neurodegeneration and cancer, and interest in modulating autophagy as a potential treatment for these diseases has grown rapidly.

In cell biology, a phagosome is a vesicle formed around a particle engulfed by a phagocyte via phagocytosis. Professional phagocytes include macrophages, neutrophils, and dendritic cells (DCs).
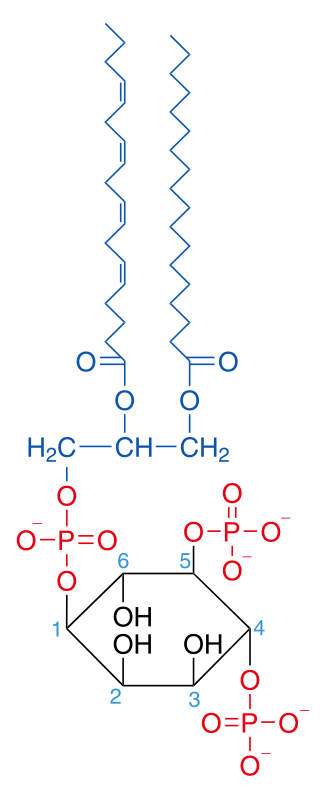
Phosphatidylinositol 4,5-bisphosphate or PtdIns(4,5)P2, also known simply as PIP2 or PI(4,5)P2, is a minor phospholipid component of cell membranes. PtdIns(4,5)P2 is enriched at the plasma membrane where it is a substrate for a number of important signaling proteins. PIP2 also forms lipid clusters that sort proteins.

Vojo Deretic, is distinguished professor and chair of the Department of Molecular Genetics and Microbiology at the University of New Mexico School of Medicine. Deretic was the founding director of the Autophagy, Inflammation and Metabolism (AIM) Center of Biomedical Research Excellence. The AIM center promotes autophagy research nationally and internationally.
Non-structural Protein 6 (NSP6) is one of the two non-structural proteins that gene 11 in rotavirus encodes for alongside NSP5. It is a putative transmembrane domain protein. NSP6 is composed of six transmembrane domains and a C terminal tail. In contrast to the other rotavirus non-structural proteins, NSP6 was found to have a high rate of turnover, being completely degraded within 2 hours of synthesis. NSP6 was found to be a sequence-independent nucleic acid binding protein, with similar affinities for ssRNA and dsRNA

Autophagy protein 5 (ATG5) is a protein that, in humans, is encoded by the ATG5 gene located on chromosome 6. It is an E3 ubi autophagic cell death. ATG5 is a key protein involved in the extension of the phagophoric membrane in autophagic vesicles. It is activated by ATG7 and forms a complex with ATG12 and ATG16L1. This complex is necessary for LC3-I conjugation to PE (phosphatidylethanolamine) to form LC3-II. ATG5 can also act as a pro-apoptotic molecule targeted to the mitochondria. Under low levels of DNA damage, ATG5 can translocate to the nucleus and interact with survivin.

Microtubule-associated proteins 1A/1B light chain 3B is a protein that in humans is encoded by the MAP1LC3B gene. LC3 is a central protein in the autophagy pathway where it functions in autophagy substrate selection and autophagosome biogenesis. LC3 is the most widely used marker of autophagosomes.

Autophagy related 16 like 1 is a protein that in humans is encoded by the ATG16L1 gene. This protein is characterized as a subunit of the autophagy-related ATG12-ATG5/ATG16 complex and is essentially important for the LC3 (ATG8) lipidation and autophagosome formation. This complex localizes to the membrane and is released just before or after autophagosome completion.
Membrane contact sites (MCS) are close appositions between two organelles. Ultrastructural studies typically reveal an intermembrane distance in the order of the size of a single protein, as small as 10 nm or wider, with no clear upper limit. These zones of apposition are highly conserved in evolution. These sites are thought to be important to facilitate signalling, and they promote the passage of small molecules, including ions, lipids and reactive oxygen species. MCS are important in the function of the endoplasmic reticulum (ER), since this is the major site of lipid synthesis within cells. The ER makes close contact with many organelles, including mitochondria, Golgi, endosomes, lysosomes, peroxisomes, chloroplasts and the plasma membrane. Both mitochondria and sorting endosomes undergo major rearrangements leading to fission where they contact the ER. Sites of close apposition can also form between most of these organelles most pairwise combinations. First mentions of these contact sites can be found in papers published in the late 1950s mainly visualized using electron microscopy (EM) techniques. Copeland and Dalton described them as “highly specialized tubular form of endoplasmic reticulum in association with the mitochondria and apparently in turn, with the vascular border of the cell”.

Autophagy-related protein 8 (Atg8) is a ubiquitin-like protein required for the formation of autophagosomal membranes. The transient conjugation of Atg8 to the autophagosomal membrane through a ubiquitin-like conjugation system is essential for autophagy in eukaryotes. Even though there are homologues in animals, this article mainly focuses on its role in lower eukaryotes such as Saccharomyces cerevisiae.

Sec14 is a cytosolic protein found in yeast which plays a role in the regulation of several cellular functions, specifically those related to intracellular transport. Encoded by the Sec14 gene, Sec14p may transport phosphatidylinositol and phosphatidylcholine produced in the endoplasmic reticulum and the Golgi body to other cellular membranes. Additionally, Sec14p potentially plays a role in the localization of lipid raft proteins. Sec14p is an essential gene in yeast, and is homologous in function to phosphatidylinositol transfer protein in mammals. A conditional mutant with non-functional Sec14p presents with Berkeley bodies and deficiencies in protein secretion.
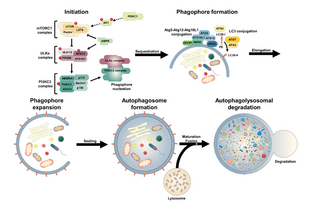
An autophagosome is a spherical structure with double layer membranes. It is the key structure in macroautophagy, the intracellular degradation system for cytoplasmic contents. After formation, autophagosomes deliver cytoplasmic components to the lysosomes. The outer membrane of an autophagosome fuses with a lysosome to form an autolysosome. The lysosome's hydrolases degrade the autophagosome-delivered contents and its inner membrane.
Chaperone-assisted selective autophagy is a cellular process for the selective, ubiquitin-dependent degradation of chaperone-bound proteins in lysosomes.
Microautophagy is one of the three common forms of autophagic pathway, but unlike macroautophagy and chaperone-mediated autophagy, it is mediated—in mammals by lysosomal action or in plants and fungi by vacuolar action—by direct engulfment of the cytoplasmic cargo. Cytoplasmic material is trapped in the lysosome/vacuole by a random process of membrane invagination.

Intracellular transport is the movement of vesicles and substances within a cell. Intracellular transport is required for maintaining homeostasis within the cell by responding to physiological signals. Proteins synthesized in the cytosol are distributed to their respective organelles, according to their specific amino acid’s sorting sequence. Eukaryotic cells transport packets of components to particular intracellular locations by attaching them to molecular motors that haul them along microtubules and actin filaments. Since intracellular transport heavily relies on microtubules for movement, the components of the cytoskeleton play a vital role in trafficking vesicles between organelles and the plasma membrane by providing mechanical support. Through this pathway, it is possible to facilitate the movement of essential molecules such as membrane‐bounded vesicles and organelles, mRNA, and chromosomes.

GRAM domain containing 1A also known as Aster-A is a protein that is encoded by the GRAMD1A gene. It contains a transmembrane region, a GRAM domain and a VASt domain that can bind cholesterol. GRAMD1A has four paralogs: GRAMD1B and GRAMD1C and two without VASt domains, GRAMD2A and GRAMD2B. These proteins are mammalian representatives of the yeast lipid transfer proteins anchored at a membrane contact site (LAM) family.
Phosphatidylinositol 3-phosphate-binding protein 2 (Pib2) is a yeast protein involved in the regulation of TORC1 signaling and lysosomal membrane permeabilization. It is essential for the reactivation of TORC1 following exposure to rapamycin or nutrient starvation.

Rubicon is a protein that in humans is encoded by the RUBCN gene. Rubicon is one of the few known negative regulators of autophagy, a cellular process that degrades unnecessary or damaged cellular components. Rubicon is recruited to its sites of action through interaction with the small GTPase Rab7, and impairs the autophagosome-lysosome fusion step of autophagy through inhibition of PI3KC3-C2.
Atg8ylation is a process of conjugation of mammalian ATG8 proteins (mATG8s) to proteins or membranes. The process is akin to the ubiquitylation of diverse substrates by ubiquitin. There are six principal mATG8s: LC3A, LC3B, LC3C, GABARAP, GABARAPL1 and GABARAPL2. Together, they comprise a sub-class of ubiquitin-like molecules characterized by two N-terminal α-helices added to the ubiquitin core, which serve a dual role of forming a docking site for interacting proteins containing ATG8-interaction motifs and enhancing mATG8’s affinity for membranes.













