
Bacillus cereus is a Gram-positive rod-shaped bacterium commonly found in soil, food, and marine sponges. The specific name, cereus, meaning "waxy" in Latin, refers to the appearance of colonies grown on blood agar. Some strains are harmful to humans and cause foodborne illness due to their spore-forming nature, while other strains can be beneficial as probiotics for animals, and even exhibit mutualism with certain plants. B. cereus bacteria may be aerobes or facultative anaerobes, and like other members of the genus Bacillus, can produce protective endospores. They have a wide range of virulence factors, including phospholipase C, cereulide, sphingomyelinase, metalloproteases, and cytotoxin K, many of which are regulated via quorum sensing. B. cereus strains exhibit flagellar motility.
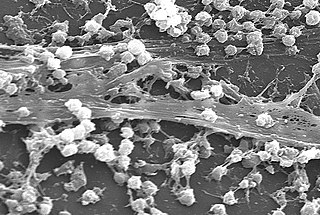
A biofilm is a syntrophic community of microorganisms in which cells stick to each other and often also to a surface. These adherent cells become embedded within a slimy extracellular matrix that is composed of extracellular polymeric substances (EPSs). The cells within the biofilm produce the EPS components, which are typically a polymeric combination of extracellular polysaccharides, proteins, lipids and DNA. Because they have a three-dimensional structure and represent a community lifestyle for microorganisms, they have been metaphorically described as "cities for microbes".
In biology, quorum sensing or quorum signaling (QS) is the process of cell-to-cell communication that allows bacteria to detect and respond to cell population density by gene regulation, typically as a means of acclimating to environmental disadvantages.

Pseudomonas aeruginosa is a common encapsulated, Gram-negative, aerobic–facultatively anaerobic, rod-shaped bacterium that can cause disease in plants and animals, including humans. A species of considerable medical importance, P. aeruginosa is a multidrug resistant pathogen recognized for its ubiquity, its intrinsically advanced antibiotic resistance mechanisms, and its association with serious illnesses – hospital-acquired infections such as ventilator-associated pneumonia and various sepsis syndromes. P. aeruginosa is able to selectively inhibit various antibiotics from penetrating its outer membrane - and has high resistance to several antibiotics. According to the World Health Organization P. aeruginosa poses one of the greatest threats to humans in terms of antibiotic resistance.
Virulence factors are cellular structures, molecules and regulatory systems that enable microbial pathogens to achieve the following:

The trp operon is a group of genes that are transcribed together, encoding the enzymes that produce the amino acid tryptophan in bacteria. The trp operon was first characterized in Escherichia coli, and it has since been discovered in many other bacteria. The operon is regulated so that, when tryptophan is present in the environment, the genes for tryptophan synthesis are repressed.
The gene rpoS encodes the sigma factor sigma-38, a 37.8 kD protein in Escherichia coli. Sigma factors are proteins that regulate transcription in bacteria. Sigma factors can be activated in response to different environmental conditions. rpoS is transcribed in late exponential phase, and RpoS is the primary regulator of stationary phase genes. RpoS is a central regulator of the general stress response and operates in both a retroactive and a proactive manner: it not only allows the cell to survive environmental challenges, but it also prepares the cell for subsequent stresses (cross-protection). The transcriptional regulator CsgD is central to biofilm formation, controlling the expression of the curli structural and export proteins, and the diguanylate cyclase, adrA, which indirectly activates cellulose production. The rpoS gene most likely originated in the gammaproteobacteria.

fis is an E. coli gene encoding the Fis protein. The regulation of this gene is more complex than most other genes in the E. coli genome, as Fis is an important protein which regulates expression of other genes. It is supposed that fis is regulated by H-NS, IHF and CRP. It also regulates its own expression (autoregulation). Fis is one of the most abundant DNA binding proteins in Escherichia coli under nutrient-rich growth conditions.

The MicA RNA is a small non-coding RNA that was discovered in E. coli during a large scale screen. Expression of SraD is highly abundant in stationary phase, but low levels could be detected in exponentially growing cells as well.

(p)ppGpp, guanosine pentaphosphate and tetraphosphate, also known as the "magic spot" nucleotides, are alarmones involved in the stringent response in bacteria that cause the inhibition of RNA synthesis when there is a shortage of amino acids. This inhibition by (p)ppGpp decreases translation in the cell, conserving amino acids present. Furthermore, ppGpp and pppGpp cause the up-regulation of many other genes involved in stress response such as the genes for amino acid uptake and biosynthesis. (p)ppGpp is also conserved in plants, where it is known to play a role in regulating growth and developmental processes.
In biology, an autoinducer is a signaling molecule that enables detection and response to changes in the population density of bacterial cells. Synthesized when a bacterium reproduces, autoinducers pass outside the bacterium and into the surrounding medium. They are a key component of the phenomenon of quorum sensing: as the density of quorum-sensing bacterial cells increases, so does the concentration of the autoinducer. A bacterium’s detection of an autoinducer above some minimum threshold triggers altered gene expression.
Osmoprotectants or compatible solutes are small organic molecules with neutral charge and low toxicity at high concentrations that act as osmolytes and help organisms survive extreme osmotic stress. Osmoprotectants can be placed in three chemical classes: betaines and associated molecules, sugars and polyols, and amino acids. These molecules accumulate in cells and balance the osmotic difference between the cell's surroundings and the cytosol. In plants, their accumulation can increase survival during stresses such as drought. In extreme cases, such as in bdelloid rotifers, tardigrades, brine shrimp, and nematodes, these molecules can allow cells to survive being completely dried out and let them enter a state of suspended animation called cryptobiosis.

The rsmX gene is part of the Rsm/Csr family of non-coding RNAs (ncRNAs). Members of the Rsm/Csr family are present in a diverse range of bacteria, including Escherichia coli, Erwinia, Salmonella, Vibrio and Pseudomonas. These ncRNAs act by sequestering translational repressor proteins, called RsmA, activating expression of downstream genes that would normally be blocked by the repressors. Sequestering of target proteins is dependent upon exposed GGA motifs in the stem loops of the ncRNAs. Typically, the activated genes are involved in secondary metabolism, biofilm formation and motility.
Bacterial small RNAs are small RNAs produced by bacteria; they are 50- to 500-nucleotide non-coding RNA molecules, highly structured and containing several stem-loops. Numerous sRNAs have been identified using both computational analysis and laboratory-based techniques such as Northern blotting, microarrays and RNA-Seq in a number of bacterial species including Escherichia coli, the model pathogen Salmonella, the nitrogen-fixing alphaproteobacterium Sinorhizobium meliloti, marine cyanobacteria, Francisella tularensis, Streptococcus pyogenes, the pathogen Staphylococcus aureus, and the plant pathogen Xanthomonas oryzae pathovar oryzae. Bacterial sRNAs affect how genes are expressed within bacterial cells via interaction with mRNA or protein, and thus can affect a variety of bacterial functions like metabolism, virulence, environmental stress response, and structure.
Rsa RNAs are non-coding RNAs found in the bacterium Staphylococcus aureus. The shared name comes from their discovery, and does not imply homology. Bioinformatics scans identified the 16 Rsa RNA families named RsaA-K and RsaOA-OG. Others, RsaOH-OX, were found thanks to an RNomic approach. Although the RNAs showed varying expression patterns, many of the newly discovered RNAs were shown to be Hfq-independent and most carried a C-rich motif (UCCC).

In molecular biology, the LuxR-type DNA-binding HTH domain is a DNA-binding, helix-turn-helix (HTH) domain of about 65 amino acids. It is present in transcription regulators of the LuxR/FixJ family of response regulators. The domain is named after Vibrio fischeri luxR, a transcriptional activator for quorum-sensing control of luminescence. LuxR-type HTH domain proteins occur in a variety of organisms. The DNA-binding HTH domain is usually located in the C-terminal region of the protein; the N-terminal region often containing an autoinducer-binding domain or a response regulatory domain. Most luxR-type regulators act as transcription activators, but some can be repressors or have a dual role for different sites. LuxR-type HTH regulators control a wide variety of activities in various biological processes.

Rhamnolipids are a class of glycolipid produced by Pseudomonas aeruginosa, amongst other organisms, frequently cited as bacterial surfactants. They have a glycosyl head group, in this case a rhamnose moiety, and a 3-(hydroxyalkanoyloxy)alkanoic acid (HAA) fatty acid tail, such as 3-hydroxydecanoic acid.
Bacterial morphological plasticity refers to changes in the shape and size that bacterial cells undergo when they encounter stressful environments. Although bacteria have evolved complex molecular strategies to maintain their shape, many are able to alter their shape as a survival strategy in response to protist predators, antibiotics, the immune response, and other threats.

Bacterial secretion systems are protein complexes present on the cell membranes of bacteria for secretion of substances. Specifically, they are the cellular devices used by pathogenic bacteria to secrete their virulence factors to invade the host cells. They can be classified into different types based on their specific structure, composition and activity. Generally, proteins can be secreted through two different processes. One process is a one-step mechanism in which proteins from the cytoplasm of bacteria are transported and delivered directly through the cell membrane into the host cell. Another involves a two-step activity in which the proteins are first transported out of the inner cell membrane, then deposited in the periplasm, and finally through the outer cell membrane into the host cell.
Diffusible signal factor (DSF) is found in several gram-negative bacteria and play a role in the formation of biofilms, motility, virulence, and antibiotic resistance. Xanthomonas campestris was the first bacteria known to have DSF. The synthesis of the DSF can be seen in rpfF and rpfB enzymes. An understanding of the DSF signaling mechanism could lead to further disease control.











