Related Research Articles

Plasmodium falciparum is a unicellular protozoan parasite of humans, and the deadliest species of Plasmodium that causes malaria in humans. The parasite is transmitted through the bite of a female Anopheles mosquito and causes the disease's most dangerous form, falciparum malaria. It is responsible for around 50% of all malaria cases. P. falciparum is therefore regarded as the deadliest parasite in humans. It is also associated with the development of blood cancer and is classified as a Group 2A (probable) carcinogen.

CD36, also known as platelet glycoprotein 4, fatty acid translocase (FAT), scavenger receptor class B member 3 (SCARB3), and glycoproteins 88 (GP88), IIIb (GPIIIB), or IV (GPIV) is a protein that in humans is encoded by the CD36 gene. The CD36 antigen is an integral membrane protein found on the surface of many cell types in vertebrate animals. It imports fatty acids inside cells and is a member of the class B scavenger receptor family of cell surface proteins. CD36 binds many ligands including collagen, thrombospondin, erythrocytes parasitized with Plasmodium falciparum, oxidized low density lipoprotein, native lipoproteins, oxidized phospholipids, and long-chain fatty acids.
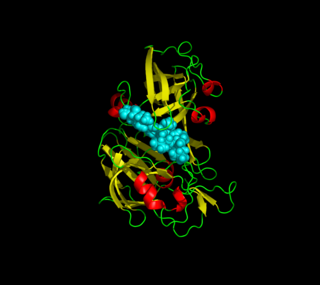
Plasmepsins are a class of at least 10 enzymes produced by the Plasmodium falciparum parasite. There are ten different isoforms of these proteins and ten genes coding them respectively in Plasmodium. It has been suggested that the plasmepsin family is smaller in other human Plasmodium species. Expression of Plm I, II, IV, V, IX, X and HAP occurs in the erythrocytic cycle, and expression of Plm VI, VII, VIII, occurs in the exoerythrocytic cycle. Through their haemoglobin-degrading activity, they are an important cause of symptoms in malaria sufferers. Consequently, this family of enzymes is a potential target for antimalarial drugs.

Plasmodium berghei is a species in the genus Plasmodium subgenus Vinckeia.

Tetracycline-controlled transcriptional activation is a method of inducible gene expression where transcription is reversibly turned on or off in the presence of the antibiotic tetracycline or one of its derivatives.
Site-specific recombinase technologies are genome engineering tools that depend on recombinase enzymes to replace targeted sections of DNA.
Antigenic variation or antigenic alteration refers to the mechanism by which an infectious agent such as a protozoan, bacterium or virus alters the proteins or carbohydrates on its surface and thus avoids a host immune response, making it one of the mechanisms of antigenic escape. It is related to phase variation. Antigenic variation not only enables the pathogen to avoid the immune response in its current host, but also allows re-infection of previously infected hosts. Immunity to re-infection is based on recognition of the antigens carried by the pathogen, which are "remembered" by the acquired immune response. If the pathogen's dominant antigen can be altered, the pathogen can then evade the host's acquired immune system. Antigenic variation can occur by altering a variety of surface molecules including proteins and carbohydrates. Antigenic variation can result from gene conversion, site-specific DNA inversions, hypermutation, or recombination of sequence cassettes. The result is that even a clonal population of pathogens expresses a heterogeneous phenotype. Many of the proteins known to show antigenic or phase variation are related to virulence.

A malaria vaccine is a vaccine that is used to prevent malaria. The only approved vaccine, as of 2021, is RTS,S, known by the brand name Mosquirix. In October 2021, the WHO for the first time recommended the large-scale use of a malaria vaccine for children living in areas with moderate-to-high malaria transmission. Four injections are required for full protection.

Macrophage migration inhibitory factor (MIF), also known as glycosylation-inhibiting factor (GIF), L-dopachrome isomerase, or phenylpyruvate tautomerase is a protein that in humans is encoded by the MIF gene. MIF is an important regulator of innate immunity. The MIF protein superfamily also includes a second member with functionally related properties, the D-dopachrome tautomerase (D-DT). CD74 is a surface receptor for MIF.

Haemozoin is a disposal product formed from the digestion of blood by some blood-feeding parasites. These hematophagous organisms such as malaria parasites, Rhodnius and Schistosoma digest haemoglobin and release high quantities of free heme, which is the non-protein component of haemoglobin. Heme is a prosthetic group consisting of an iron atom contained in the center of a heterocyclic porphyrin ring. Free heme is toxic to cells, so the parasites convert it into an insoluble crystalline form called hemozoin. In malaria parasites, hemozoin is often called malaria pigment.
Human genetic resistance to malaria refers to inherited changes in the DNA of humans which increase resistance to malaria and result in increased survival of individuals with those genetic changes. The existence of these genotypes is likely due to evolutionary pressure exerted by parasites of the genus Plasmodium which cause malaria. Since malaria infects red blood cells, these genetic changes are most common alterations to molecules essential for red blood cell function, such as hemoglobin or other cellular proteins or enzymes of red blood cells. These alterations generally protect red blood cells from invasion by Plasmodium parasites or replication of parasites within the red blood cell.
The N-end rule is a rule that governs the rate of protein degradation through recognition of the N-terminal residue of proteins. The rule states that the N-terminal amino acid of a protein determines its half-life. The rule applies to both eukaryotic and prokaryotic organisms, but with different strength, rules, and outcome. In eukaryotic cells, these N-terminal residues are recognized and targeted by ubiquitin ligases, mediating ubiquitination thereby marking the protein for degradation. The rule was initially discovered by Alexander Varshavsky and co-workers in 1986. However, only rough estimations of protein half-life can be deduced from this 'rule', as N-terminal amino acid modification can lead to variability and anomalies, whilst amino acid impact can also change from organism to organism. Other degradation signals, known as degrons, can also be found in sequence.
Pregnancy-associated malaria (PAM) or placental malaria is a presentation of the common illness that is particularly life-threatening to both mother and developing fetus. PAM is caused primarily by infection with Plasmodium falciparum, the most dangerous of the four species of malaria-causing parasites that infect humans. During pregnancy, a woman faces a much higher risk of contracting malaria and of associated complications. Prevention and treatment of malaria are essential components of prenatal care in areas where the parasite is endemic – tropical and subtropical geographic areas. Placental malaria has also been demonstrated to occur in animal models, including in rodent and non-human primate models.

In molecular biology, Duffy binding proteins are found in Plasmodium. Plasmodium vivax and Plasmodium knowlesi merozoites invade Homo sapiens erythrocytes that express Duffy blood group surface determinants. The Duffy receptor family is localised in micronemes, an organelle found in all organisms of the phylum Apicomplexa.
The AP2-Coincident Domain mainly at the Carboxy-terminus, or ACDC domain, is a protein domain that occurs in proteins from apicomplexan parasites.

Chemically Induced Dimerization (CID) is a biological mechanism in which two proteins bind only in the presence of a certain small molecule, enzyme or other dimerizing agent. Genetically engineered CID systems are used in biological research to control protein localization, to manipulate signalling pathways and to induce protein activation.
Circumsporozoite protein (CSP) is a secreted protein of the sporozoite stage of the malaria parasite and is the antigenic target of RTS,S, a pre-erythrocytic malaria vaccine currently undergoing clinical trials. The amino-acid sequence of CSP consists of an immunodominant central repeat region flanked by conserved motifs at the N- and C- termini that are implicated in protein processing as the parasite travels from the mosquito to the mammalian vector. CSP was discovered by Dame et al 1984 in P. gallinaceum.
Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) is a family of proteins present on the membrane surface of red blood cells that are infected by the malarial parasite Plasmodium falciparum. PfEMP1 is synthesized during the parasite's blood stage inside the RBC, during which the clinical symptoms of falciparum malaria are manifested. Acting as both an antigen and adhesion protein, it is thought to play a key role in the high level of virulence associated with P. falciparum. It was discovered in 1984 when it was reported that infected RBCs had unusually large-sized cell membrane proteins, and these proteins had antibody-binding (antigenic) properties. An elusive protein, its chemical structure and molecular properties were revealed only after a decade, in 1995. It is now established that there is not one but a large family of PfEMP1 proteins, genetically regulated (encoded) by a group of about 60 genes called var. Each P. falciparum is able to switch on and off specific var genes to produce a functionally different protein, thereby evading the host's immune system. RBCs carrying PfEMP1 on their surface stick to endothelial cells, which facilitates further binding with uninfected RBCs, ultimately helping the parasite to both spread to other RBCs as well as bringing about the fatal symptoms of P. falciparum malaria.

The Plasmodium helical interspersed subtelomeric proteins (PHIST) or ring-infected erythrocyte surface antigens (RESA) are a family of protein domains found in the malaria-causing Plasmodium species. It was initially identified as a short four-helical conserved region in the single-domain export proteins, but the identification of this part associated with a DnaJ domain in P. falciparum RESA has led to its reclassification as the RESA N-terminal domain. This domain has been classified into three subfamilies, PHISTa, PHISTb, and PHISTc.
Reticulocyte binding protein homologs (RHs) are a superfamily of proteins found in Plasmodium responsible for cell invasion. Together with the family of erythrocyte binding-like proteins (EBLs) they make up the two families of invasion proteins universal to Plasmodium. The two families function cooperatively.
References
- ↑ Meissner, M; Krejany, E; Gilson, PR; De Koning-Ward, TF; Soldati, D; Crabb, BS (2005). "Tetracycline analogue-regulated transgene expression in Plasmodium falciparum blood stages using Toxoplasma gondii transactivators". PNAS . 102 (8): 2980–2985. Bibcode:2005PNAS..102.2980M. doi: 10.1073/pnas.0500112102 . PMC 548799 . PMID 15710888.
- ↑ Nkrumah, LJ; Muhle, RA; Moura, PA; Ghosh, P; Hatfull, GF; Jacobs Jr, WR; Fidock, DA (2006). "Efficient site-specific integration in Plasmodium falciparum chromosomes mediated by mycobacteriophage Bxb1 integrase". Nature Methods . 3 (8): 615–621. doi:10.1038/nmeth904. PMC 2943413 . PMID 16862136.
- ↑ Carvalho, TG; Thiberge, S; Sakamoto, H; Ménard, R (2004). "Conditional mutagenesis using site-specific recombination in Plasmodium berghei transactivators". PNAS. 101 (41): 14931–14936. doi: 10.1073/pnas.0404416101 . PMC 522007 . PMID 15465918.
- ↑ Balu, B; Shoue, DA; Fraser Jr, MJ; Adams, JH (2005). "High-efficiency transformation of Plasmodium falciparum by the lepidopteran transposable element piggyBac". PNAS. 102 (45): 16391–16396. Bibcode:2005PNAS..10216391B. doi: 10.1073/pnas.0504679102 . PMC 1275597 . PMID 16260745.
- ↑ Agop-Nersesian (2008). "Functional expression of ribozymes in Apicomplexa: Towards exogenous control of gene expression by inducible RNA-cleavage". International Journal for Parasitology . 38 (6): 673–81. doi:10.1016/j.ijpara.2007.10.015. PMID 18062972.
- ↑ Armstrong, CM; Goldberg, DE (2007). "An FKBP destabilization domain modulates protein levels in Plasmodium falciparum". Nature Methods. 4 (12): 1007–1009. doi:10.1038/nmeth1132. PMID 17994030. S2CID 3314981.
- ↑ Muralidharan, V; Goldberg, DE (2011). "Asparagine repeat function in a Plasmodium falciparum protein assessed via a regulatable fluorescent affinity tag". PNAS. 108 (11): 4411–4416. Bibcode:2011PNAS..108.4411M. doi: 10.1073/pnas.1018449108 . PMC 3060247 . PMID 21368162.