
Meiosis is a special type of cell division of germ cells in sexually-reproducing organisms used to produce the gametes, such as sperm or egg cells. It involves two rounds of division that ultimately result in four cells with only one copy of each chromosome (haploid). Additionally, prior to the division, genetic material from the paternal and maternal copies of each chromosome is crossed over, creating new combinations of code on each chromosome. Later on, during fertilisation, the haploid cells produced by meiosis from a male and female will fuse to create a cell with two copies of each chromosome again, the zygote.

In cell biology, mitosis is a part of the cell cycle in which replicated chromosomes are separated into two new nuclei. Cell division gives rise to genetically identical cells in which the total number of chromosomes is maintained. Therefore, mitosis is also known as equational division. In general, mitosis is preceded by the S stage of interphase and is often followed by telophase and cytokinesis; which divides the cytoplasm, organelles and cell membrane of one cell into two new cells containing roughly equal shares of these cellular components. The different stages of Mitosis altogether define the mitotic (M) phase of an animal cell cycle—the division of the mother cell into two daughter cells genetically identical to each other.

{{distinguish|all is ronng
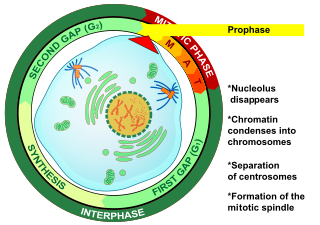
Prophase (from the Greek πρό, "before" and φάσις, "stage") is the first stage of cell division in both mitosis and meiosis. Beginning after interphase, DNA has already been replicated when the cell enters prophase. The main occurrences in prophase are the condensation of the chromatin reticulum and the disappearance of the nucleolus.

Telophase is the final stage in both meiosis and mitosis in a eukaryotic cell. During telophase, the effects of prophase and prometaphase are reversed. As chromosomes reach the cell poles, a nuclear envelope is re-assembled around each set of chromatids, the nucleoli reappear, and chromosomes begin to decondense back into the expanded chromatin that is present during interphase. The mitotic spindle is disassembled and remaining spindle microtubules are depolymerized. Telophase accounts for approximately 2% of the cell cycle's duration.

Nondisjunction is the failure of homologous chromosomes or sister chromatids to separate properly during cell division. There are three forms of nondisjunction: failure of a pair of homologous chromosomes to separate in meiosis I, failure of sister chromatids to separate during meiosis II, and failure of sister chromatids to separate during mitosis. Nondisjunction results in daughter cells with abnormal chromosome numbers (aneuploidy).

The spindle checkpoint, also known as the metaphase-to-anaphase transition, the spindle assembly checkpoint (SAC), or the mitotic checkpoint, is a cell cycle checkpoint during mitosis or meiosis that prevents the separation of the duplicated chromosomes (anaphase) until each chromosome is properly attached to the spindle. To achieve proper segregation, the two kinetochores on the sister chromatids must be attached to opposite spindle poles. Only this pattern of attachment will ensure that each daughter cell receives one copy of the chromosome. The defining biochemical feature of this checkpoint is the stimulation of the anaphase-promoting complex by M-phase cyclin-CDK complexes, which in turn causes the proteolytic destruction of cyclins and proteins that hold the sister chromatids together.

A kinetochore is a disc-shaped protein structure associated with duplicated chromatids in eukaryotic cells where the spindle fibers attach during cell division to pull sister chromatids apart. The kinetochore assembles on the centromere and links the chromosome to microtubule polymers from the mitotic spindle during mitosis and meiosis. The term kinetochore was first used in a footnote in a 1934 Cytology book by Lester W. Sharp and commonly accepted in 1936. Sharp's footnote reads: "The convenient term kinetochore has been suggested to the author by J. A. Moore", likely referring to John Alexander Moore who had joined Columbia University as a freshman in 1932.
Mad2 is an essential spindle checkpoint protein. The spindle checkpoint system is a regulatory system that restrains progression through the metaphase-to-anaphase transition. The Mad2 gene was first identified in the yeast S. cerevisiae in a screen for genes which when mutated would confer sensitivity to microtubule poisons. The human orthologues of Mad2 were first cloned in a search for human cDNAs that would rescue the microtubule poison-sensitivity of a yeast strain in which a kinetochore binding protein was missing. The protein was shown to be present at unattached kinetochores and antibody inhibition studies demonstrated it was essential to execute a block in the metaphase-to-anaphase transition in response to the microtubule poison nocodazole. Subsequent cloning of the Xenopus laevis orthologue, facilitated by the sharing of the human sequence, allowed for the characterization of the mitotic checkpoint in egg extracts.

Aurora kinase A also known as serine/threonine-protein kinase 6 is an enzyme that in humans is encoded by the AURKA gene.

Aurora B kinase is a protein that functions in the attachment of the mitotic spindle to the centromere.

Mitotic checkpoint serine/threonine-protein kinase BUB1 also known as BUB1 is an enzyme that in humans is encoded by the BUB1 gene.

Mitotic checkpoint protein BUB3 is a protein that in humans is encoded by the BUB3 gene.

Serine/threonine-protein kinase 13 is an enzyme that in humans is encoded by the AURKC gene.

HORMA domain-containing protein 1 (HORMAD1) also known as cancer/testis antigen 46 (CT46) is a protein that in humans is encoded by the HORMAD1 gene.

The meiotic recombination checkpoint monitors meiotic recombination during meiosis, and blocks the entry into metaphase I if recombination is not efficiently processed.
In cell biology, Meiomitosis is an aberrant cellular division pathway that combines normal mitosis pathways with ectopically expressed meiotic machinery resulting in genomic instability.
Cdc14 and Cdc14 are a gene and its protein product respectively. Cdc14 is found in most of the eukaryotes. Cdc14 was defined by Hartwell in his famous screen for loci that control the cell cycle of Saccharomyces cerevisiae. Cdc14 was later shown to encode a protein phosphatase. Cdc14 is dual-specificity, which means it has serine/threonine and tyrosine-directed activity. A preference for serines next to proline is reported. Many early studies, especially in the budding yeast Saccharomyces cerevisiae, demonstrated that the protein plays a key role in regulating late mitotic processes. However, more recent work in a range of systems suggests that its cellular function is more complex.

Mad1 is a non-essential protein which in yeast has a function in the spindle assembly checkpoint (SAC). This checkpoint monitors chromosome attachment to spindle microtubules and prevents cells from starting anaphase until the spindle is built up. The name Mad refers to the observation that mutant cells are mitotic arrest deficient (MAD) during microtubule depolymerization. Mad1 recruits the anaphase inhibitor Mad2 to unattached kinetochores and is essential for Mad2-Cdc20 complex formation in vivo but not in vitro. In vivo, Mad1 acts as a competitive inhibitor of the Mad2-Cdc20 complex. Mad1 is phosphorylated by Mps1 which then leads together with other activities to the formation of the mitotic checkpoint complex (MCC). Thereby it inhibits the activity of the anaphase-promoting complex/cyclosome (APC/C). Homologs of Mad1 are conserved in eukaryotes from yeast to mammals.
In molecular biology, the HORMA domain is a protein domain that has been suggested to recognise chromatin states resulting from DNA adducts, double stranded breaks or non-attachment to the spindle and act as an adaptor that recruits other proteins. Hop1 is a meiosis-specific protein, Rev7 is required for DNA damage induced mutagenesis, and MAD2 is a spindle checkpoint protein which prevents progression of the cell cycle upon detection of a defect in mitotic spindle integrity.



















