
A toroid repeat is a protein fold composed of repeating subunits, arranged in circular fashion to form a closed structure. [1] [2]

A toroid repeat is a protein fold composed of repeating subunits, arranged in circular fashion to form a closed structure. [1] [2]
In the case when the N- and C-terminal repeats lie in close physical contact in a tandem repeat domain, the result is a topologically compact, closed structure. Such domains typically display a high rotational symmetry (unlike open repeats that only have translational symmetries), and assume a wheel-like shape. Because of the limitations of this structure, the number of individual repeats is not arbitrary. In the case of WD40 repeats (perhaps the largest family of closed solenoids) the number of repeats can range from 4 to 10 (more usually between 5 and 7). [3] Kelch repeats, beta-barrels and beta-trefoil repeats are further examples for this architecture.
Closed solenoids frequently function as protein-protein interaction modules: it is possible that all repeats must be present to form the ligand-binding site if it is located at the centre or axis of the domain "wheel". The WD40 repeat is a prime example of this function. [4]
The following major sub-classes of toroid repeat proteins can be found: [1] [2]
Tandem repeats occur in DNA when a pattern of one or more nucleotides is repeated and the repetitions are directly adjacent to each other. Several protein domains also form tandem repeats within their amino acid primary structure, such as armadillo repeats. However, in proteins, perfect tandem repeats are unlikely in most in vivo proteins, and most known repeats are in proteins which have been designed.

The apoptosome is a large quaternary protein structure formed in the process of apoptosis. Its formation is triggered by the release of cytochrome c from the mitochondria in response to an internal (intrinsic) or external (extrinsic) cell death stimulus. Stimuli can vary from DNA damage and viral infection to developmental cues such as those leading to the degradation of a tadpole's tail.

A beta barrel is a beta-sheet composed of tandem repeats that twists and coils to form a closed toroidal structure in which the first strand is bonded to the last strand. Beta-strands in many beta-barrels are arranged in an antiparallel fashion. Beta barrel structures are named for resemblance to the barrels used to contain liquids. Most of them are water-soluble proteins and frequently bind hydrophobic ligands in the barrel center, as in lipocalins. Others span cell membranes and commonly found in porins. Porin-like barrel structures are encoded by as many as 2–3% of the genes in Gram-negative bacteria.

A beta helix is a tandem protein repeat structure formed by the association of parallel beta strands in a helical pattern with either two or three faces. The beta helix is a type of solenoid protein domain. The structure is stabilized by inter-strand hydrogen bonds, protein-protein interactions, and sometimes bound metal ions. Both left- and right-handed beta helices have been identified. Double stranded beta-helices are also very common features of proteins and are generally synonymous with jelly roll folds.

In structural biology, a beta-propeller is a type of all-β protein architecture characterized by 4 to 8 highly symmetrical blade-shaped beta sheets arranged toroidally around a central axis. Together the beta-sheets form a funnel-like active site.

The TIM barrel is a conserved protein fold consisting of eight α-helices and eight parallel β-strands that alternate along the peptide backbone. The structure is named after triosephosphate isomerase, a conserved metabolic enzyme. TIM barrels are ubiquitous, with approximately 10% of all enzymes adopting this fold. Further, 5 of 7 enzyme commission (EC) enzyme classes include TIM barrel proteins. The TIM barrel fold is evolutionarily ancient, with many of its members possessing little similarity today, instead falling within the twilight zone of sequence similarity.
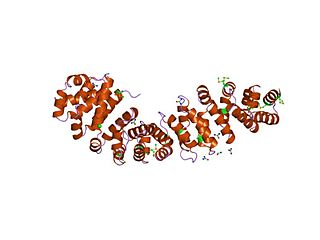
An armadillo repeat is the name of a characteristic, repetitive amino acid sequence of about 40 residues in length that is found in many proteins. Proteins that contain armadillo repeats typically contain several tandemly repeated copies. Each armadillo repeat is composed of a pair of alpha helices that form a hairpin structure. Multiple copies of the repeat form what is known as an alpha solenoid structure.

An alpha solenoid is a protein fold composed of repeating alpha helix subunits, commonly helix-turn-helix motifs, arranged in antiparallel fashion to form a superhelix. Alpha solenoids are known for their flexibility and plasticity. Like beta propellers, alpha solenoids are a form of solenoid protein domain commonly found in the proteins comprising the nuclear pore complex. They are also common in membrane coat proteins known as coatomers, such as clathrin, and in regulatory proteins that form extensive protein-protein interactions with their binding partners. Examples of alpha solenoid structures binding RNA and lipids have also been described.

A leucine-rich repeat (LRR) is a protein structural motif that forms an α/β horseshoe fold. It is composed of repeating 20–30 amino acid stretches that are unusually rich in the hydrophobic amino acid leucine. These tandem repeats commonly fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Typically, each repeat unit has beta strand-turn-alpha helix structure, and the assembled domain, composed of many such repeats, has a horseshoe shape with an interior parallel beta sheet and an exterior array of helices. One face of the beta sheet and one side of the helix array are exposed to solvent and are therefore dominated by hydrophilic residues. The region between the helices and sheets is the protein's hydrophobic core and is tightly sterically packed with leucine residues.

Kelch proteins are a widespread group of proteins that contain multiple Kelch motifs. The kelch domain generally occurs as a set of five to seven kelch tandem repeats that form a β-propeller tertiary structure. Kelch-repeat β-propellers are generally involved in protein–protein interactions, though the large diversity of domain architectures and limited sequence identity between kelch motifs make characterisation of the kelch superfamily difficult.

The WD40 repeat is a short structural motif of approximately 40 amino acids, often terminating in a tryptophan-aspartic acid (W-D) dipeptide. Tandem copies of these repeats typically fold together to form a type of circular solenoid protein domain called the WD40 domain.
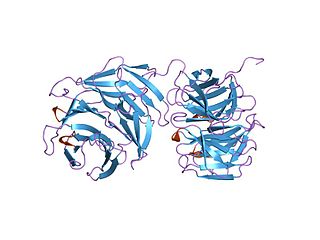
The NHL repeat, named after ncl-1, HT2A and lin-41, is an amino acid sequence found largely in a large number of eukaryotic and prokaryotic proteins. For example, the repeat is found in a variety of enzymes of the copper type II, ascorbate-dependent monooxygenase family which catalyse the C-terminus alpha-amidation of biological peptides. In many it occurs in tandem arrays, for example in the RING finger beta-box, coiled-coil (RBCC) eukaryotic growth regulators. The arthropod 'Brain Tumor' protein is one such growth regulator that contains a 6-bladed NHL-repeat beta-propeller.
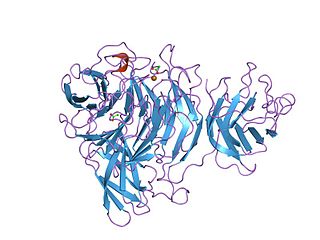
The Kelch motif is a region of protein sequence found widely in proteins from bacteria and eukaryotes. This sequence motif is composed of about 50 amino acid residues which form a structure of a four stranded beta-sheet "blade". This sequence motif is found in between five and eight tandem copies per protein which fold together to form a larger circular solenoid structure called a beta-propeller domain.

WD repeat-containing protein 72 is a protein that in humans is encoded by the WDR72 gene. WDR72 contains 7 WD40 repeats, which are predicted to form the blades of a 7 beta propeller structure.

WD repeat domain phosphoinositide-interacting protein 1 (WIPI-1), also known as Atg18 protein homolog (ATG18) and WD40 repeat protein interacting with phosphoinositides of 49 kDa, is a protein that in humans is encoded by the WIPI1 gene.

In molecular biology, the haemagglutination activity domain is a conserved protein domain found near the N terminus of a number of large, repetitive bacterial proteins, including many proteins of over 2500 amino acids. A number of the members of this family have been designated adhesins, filamentous haemagglutinins, haem/haemopexin-binding protein, etc. Members generally have a signal sequence, then an intervening region, then the region described in this entry. Following this region, proteins typically have regions rich in repeats but may show no homology between the repeats of one member and the repeats of another. This domain is suggested to be a carbohydrate-dependent haemagglutination activity site.
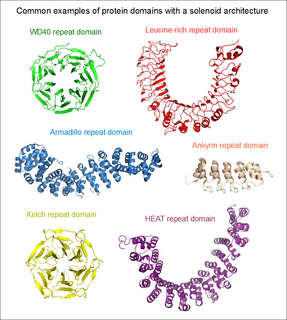
Solenoid protein domains are a highly modular type of protein domain. They consist of a chain of nearly identical folds, often simply called tandem repeats. They are extremely common among all types of proteins, though exact figures are unknown.

An array of protein tandem repeats is defined as several adjacent copies having the same or similar sequence motifs. These periodic sequences are generated by internal duplications in both coding and non-coding genomic sequences. Repetitive units of protein tandem repeats are considerably diverse, ranging from the repetition of a single amino acid to domains of 100 or more residues.
A beta solenoid is a protein fold composed of repeating beta strands subunits, arranged in antiparallel fashion to form a superhelix.