
In structural biology, a beta-propeller (β-propeller) is a type of all-β protein architecture characterized by 4 to 8 highly symmetrical blade-shaped beta sheets arranged toroidally around a central axis. Together the beta-sheets form a funnel-like active site.
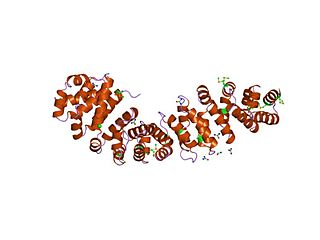
An armadillo repeat is a characteristic, repetitive amino acid sequence of about 42 residues in length that is found in many proteins. Proteins that contain armadillo repeats typically contain several tandemly repeated copies. Each armadillo repeat is composed of a pair of alpha helices that form a hairpin structure. Multiple copies of the repeat form what is known as an alpha solenoid structure.

Sialyltransferases are enzymes that transfer sialic acid to nascent oligosaccharide. Each sialyltransferase is specific for a particular sugar substrate. Sialyltransferases add sialic acid to the terminal portions of the sialylated glycolipids (gangliosides) or to the N- or O-linked sugar chains of glycoproteins.
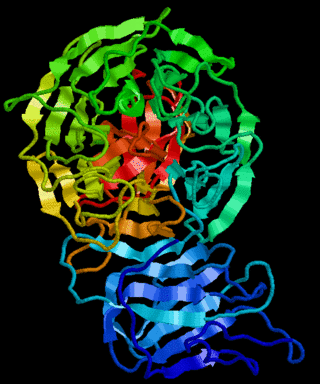
Kelch proteins are a widespread group of proteins that contain multiple Kelch motifs. The kelch domain generally occurs as a set of five to seven kelch tandem repeats that form a β-propeller tertiary structure. Kelch-repeat β-propellers are generally involved in protein–protein interactions, though the large diversity of domain architectures and limited sequence identity between kelch motifs make characterisation of the kelch superfamily difficult.

Filamin A, alpha (FLNA) is a protein that in humans is encoded by the FLNA gene.

Coronin-1A is a protein that in humans is encoded by the CORO1A gene. It has been implicated in both T-cell mediated immunity and mitochondrial apoptosis. In a recent genome-wide longevity study, its expression levels were found to be negatively associated both with age at the time of blood sample and the survival time after blood draw.

CMP-sialic acid transporter is a protein that in humans is encoded by the SLC35A1 gene.

ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4, also known as sialyltransferase 3C (SIAT3-C) or sialyltransferase 7D (SIAT7-D) is a sialyltransferase enzyme that in humans is encoded by the ST6GALNAC4 gene.

Kelch-like protein 20 is a protein that in humans is encoded by the KLHL20 gene.
Coronin is an actin binding protein which also interacts with microtubules and in some cell types is associated with phagocytosis. Coronin proteins are expressed in a large number of eukaryotic organisms from yeast to humans.
YWTD repeats are four-stranded beta-propeller repeats found in low-density lipoprotein receptors (LDLR). The six YWTD repeats together fold into a six-bladed beta-propeller. Each blade of the propeller consists of four antiparallel beta-strands; the innermost strand of each blade is labeled 1 and the outermost strand, 4. The sequence repeats are offset with respect to the blades of the propeller, such that any given 40-residue YWTD repeat spans strands 24 of one propeller blade and strand 1 of the subsequent blade. This offset ensures circularization of the propeller because the last strand of the final sequence repeat acts as an innermost strand 1 of the blade that harbors strands 24 from the first sequence repeat. The repeat is found in a variety of proteins that include, vitellogenin receptor from Drosophila melanogaster, low-density lipoprotein (LDL) receptor, preproepidermal growth factor, and nidogen (entactin).

The BTB/POZ domain is a structural domain found in proteins across the domain Eukarya. Given its prevalence in eukaryotes and its absence in Archaea and bacteria, it likely arose after the origin of eukaryotes. While primarily a protein-protein interaction domain, some BTB domains have additional functionality in transcriptional regulation, cytoskeletal mobility, protein ubiquitination and degradation, and ion channel formation and operation. BTB domains have traditionally been classified by the other structural features present in the protein.
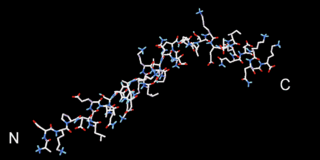
Beta thymosins are a family of proteins which have in common a sequence of about 40 amino acids similar to the small protein thymosin β4. They are found almost exclusively in multicellular animals. Thymosin β4 was originally obtained from the thymus in company with several other small proteins which although named collectively "thymosins" are now known to be structurally and genetically unrelated and present in many different animal tissues.
WH1 domain is an evolutionary conserved protein domain found on WASP proteins, which are often involved in actin polymerization.
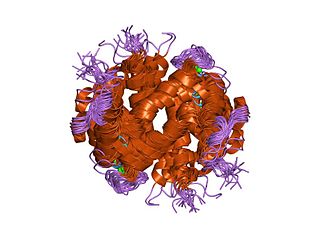
In molecular biology, the F-actin capping protein is a protein complex which binds in a calcium-independent manner to the fast-growing ends of actin filaments, thereby blocking the exchange of subunits at these ends. Unlike gelsolin and severin this protein does not sever actin filaments. The F-actin capping protein is a heterodimer composed of two unrelated subunits: alpha and beta. Neither of the subunits shows sequence similarity to other filament-capping proteins. The alpha subunit is a protein of about 268 to 286 amino acid residues and the beta subunit is approximately 280 amino acids, their sequences are well conserved in eukaryotic species.

BNR/Asp-box repeat is a repetitive sequence of amino acids contained in some proteins. Many of these proteins contain multiple BNR repeats or Asp-boxes.

Solenoid protein domains are a highly modular type of protein domain. They consist of a chain of nearly identical folds, often simply called tandem repeats. They are extremely common among all types of proteins, though exact figures are unknown.
Megf8 also known as Multiple Epidermal Growth Factor-like Domains 8, is a protein coding gene that encodes a single pass membrane protein, known to participate in developmental regulation and cellular communication. It is located on chromosome 19 at the 49th open reading frame in humans (19q13.2). There are two isoform constructs known for MEGF8, which differ by a 67 amino acid indel. The isoform 2 splice version is 2785 amino acids long, and predicted to be 296.6 kdal in mass. Isoform 1 is composed of 2845 amino acids and predicted to weigh 303.1 kdal. Using BLAST searches, orthologs were found primarily in mammals, but MEGF8 is also conserved in invertebrates and fishes, and rarely in birds, reptiles, and amphibians. A notably important paralog to multiple epidermal growth factor-like domains 8 is ATRNL1, which is also a single pass transmembrane protein, with several of the same key features and motifs as MEGF8, as indicated by Simple Modular Architecture Research Tool (SMART) which is hosted by the European Molecular Biology Laboratory located in Heidelberg, Germany. MEGF8 has been predicted to be a key player in several developmental processes, such as left-right patterning and limb formation. Currently, researchers have found MEGF8 SNP mutations to be the cause of Carpenter syndrome subtype 2.

Galactose oxidase is an enzyme that catalyzes the oxidation of D-galactose in some species of fungi.

A toroid repeat is a protein fold composed of repeating subunits, arranged in circular fashion to form a closed structure.

















