
An alpha helix is a sequence of amino acids in a protein that are twisted into a coil.

The beta sheet is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet. A β-strand is a stretch of polypeptide chain typically 3 to 10 amino acids long with backbone in an extended conformation. The supramolecular association of β-sheets has been implicated in the formation of the fibrils and protein aggregates observed in amyloidosis, Alzheimer's disease and other proteinopathies.

Collagen is the main structural protein in the extracellular matrix found in the body's various connective tissues. As the main component of connective tissue, it is the most abundant protein in mammals, making up from 25% to 35% of the whole-body protein content. Collagen consists of amino acids bound together to form a triple helix of elongated fibril known as a collagen helix. It is mostly found in connective tissue such as cartilage, bones, tendons, ligaments, and skin. Vitamin C is vital for collagen synthesis, and Vitamin E improves the production of collagen.

Protein secondary structure is the local spatial conformation of the polypeptide backbone excluding the side chains. The two most common secondary structural elements are alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein folds into its three dimensional tertiary structure.
Proline (symbol Pro or P) is an organic acid classed as a proteinogenic amino acid (used in the biosynthesis of proteins), although it does not contain the amino group -NH
2 but is rather a secondary amine. The secondary amine nitrogen is in the protonated form (NH2+) under biological conditions, while the carboxyl group is in the deprotonated −COO− form. The "side chain" from the α carbon connects to the nitrogen forming a pyrrolidine loop, classifying it as a aliphatic amino acid. It is non-essential in humans, meaning the body can synthesize it from the non-essential amino acid L-glutamate. It is encoded by all the codons starting with CC (CCU, CCC, CCA, and CCG).

(2S,4R)-4-Hydroxyproline, or L-hydroxyproline (C5H9O3N), is an amino acid, abbreviated as Hyp or O, e.g., in Protein Data Bank.
In chemistry, hydroxylation can refer to:

In biochemistry, a Ramachandran plot, originally developed in 1963 by G. N. Ramachandran, C. Ramakrishnan, and V. Sasisekharan, is a way to visualize energetically allowed regions for backbone dihedral angles ψ against φ of amino acid residues in protein structure. The figure on the left illustrates the definition of the φ and ψ backbone dihedral angles. The ω angle at the peptide bond is normally 180°, since the partial-double-bond character keeps the peptide bond planar. The figure in the top right shows the allowed φ,ψ backbone conformational regions from the Ramachandran et al. 1963 and 1968 hard-sphere calculations: full radius in solid outline, reduced radius in dashed, and relaxed tau (N-Cα-C) angle in dotted lines. Because dihedral angle values are circular and 0° is the same as 360°, the edges of the Ramachandran plot "wrap" right-to-left and bottom-to-top. For instance, the small strip of allowed values along the lower-left edge of the plot are a continuation of the large, extended-chain region at upper left.
In statistical mechanics, the Zimm–Bragg model is a helix-coil transition model that describes helix-coil transitions of macromolecules, usually polymer chains. Most models provide a reasonable approximation of the fractional helicity of a given polypeptide; the Zimm–Bragg model differs by incorporating the ease of propagation (self-replication) with respect to nucleation. It is named for co-discoverers Bruno H. Zimm and J. K. Bragg.
A polyproline helix is a type of protein secondary structure which occurs in proteins comprising repeating proline residues. A left-handed polyproline II helix is formed when sequential residues all adopt (φ,ψ) backbone dihedral angles of roughly and have trans isomers of their peptide bonds. This PPII conformation is also common in proteins and polypeptides with other amino acids apart from proline. Similarly, a more compact right-handed polyproline I helix is formed when sequential residues all adopt (φ,ψ) backbone dihedral angles of roughly and have cis isomers of their peptide bonds. Of the twenty common naturally occurring amino acids, only proline is likely to adopt the cis isomer of the peptide bond, specifically the X-Pro peptide bond; steric and electronic factors heavily favor the trans isomer in most other peptide bonds. However, peptide bonds that replace proline with another N-substituted amino acid are also likely to adopt the cis isomer.

A pi helix is a type of secondary structure found in proteins. Discovered by crystallographer Barbara Low in 1952 and once thought to be rare, short π-helices are found in 15% of known protein structures and are believed to be an evolutionary adaptation derived by the insertion of a single amino acid into an α-helix. Because such insertions are highly destabilizing, the formation of π-helices would tend to be selected against unless it provided some functional advantage to the protein. π-helices therefore are typically found near functional sites of proteins.

A 310 helix is a type of secondary structure found in proteins and polypeptides. Of the numerous protein secondary structures present, the 310-helix is the fourth most common type observed; following α-helices, β-sheets and reverse turns. 310-helices constitute nearly 10–15% of all helices in protein secondary structures, and are typically observed as extensions of α-helices found at either their N- or C- termini. Because of the α-helices tendency to consistently fold and unfold, it has been proposed that the 310-helix serves as an intermediary conformation of sorts, and provides insight into the initiation of α-helix folding.
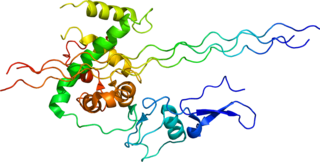
Type III Collagen is a homotrimer, or a protein composed of three identical peptide chains (monomers), each called an alpha 1 chain of type III collagen. Formally, the monomers are called collagen type III, alpha-1 chain and in humans are encoded by the COL3A1 gene. Type III collagen is one of the fibrillar collagens whose proteins have a long, inflexible, triple-helical domain.
Type I collagen is the most abundant collagen of the human body, consisting of around 90% of the body's total collagen in vertebrates. Due to this, it is also the most abundant protein type found in all vertebrates. Type I forms large, eosinophilic fibers known as collagen fibers, which make up most of the rope-like dense connective tissue in the body. Collagen I itself is created by the combination of both a proalpha1 and a proalpha2 chain created by the COL1alpha1 and COL1alpha2 genes respectively. The Col I gene itself takes up a triple-helical conformation due to its Glycine-X-Y structure, x and y being any type of amino acid. Collagen can also be found in two different isoforms, either as a homotrimer or a heterotrimer, both of which can be found during different periods of development. Heterotrimers, in particular, play an important role in wound healing, and are the dominant isoform found in the body.

Procollagen-proline dioxygenase, commonly known as prolyl hydroxylase, is a member of the class of enzymes known as alpha-ketoglutarate-dependent hydroxylases. These enzymes catalyze the incorporation of oxygen into organic substrates through a mechanism that requires alpha-Ketoglutaric acid, Fe2+, and ascorbate. This particular enzyme catalyzes the formation of (2S, 4R)-4-hydroxyproline, a compound that represents the most prevalent post-translational modification in the human proteome.

Xaa-Pro dipeptidase, also known as prolidase, is an enzyme that in humans is encoded by the PEPD gene. Prolidase is an enzyme in humans that plays a crucial role in protein metabolism and collagen recycling through the catalysis of the rate-limiting step in these chemical reactions. This enzyme is coded by the gene PEPD, located on chromosome 19. Serum prolidase activity is also currently being explored as a biomarker for diseases.
β turns are the most common form of turns—a type of non-regular secondary structure in proteins that cause a change in direction of the polypeptide chain. They are very common motifs in proteins and polypeptides. Each consists of four amino acid residues. They can be defined in two ways:
- By the possession of an intra-main-chain hydrogen bond between the CO of residue i and the NH of residue i+3;
- By having a distance of less than 7Å between the Cα atoms of residues i and i+3.

In the fields of geometry and biochemistry, a triple helix is a set of three congruent geometrical helices with the same axis, differing by a translation along the axis. This means that each of the helices keeps the same distance from the central axis. As with a single helix, a triple helix may be characterized by its pitch, diameter, and handedness. Examples of triple helices include triplex DNA, triplex RNA, the collagen helix, and collagen-like proteins.
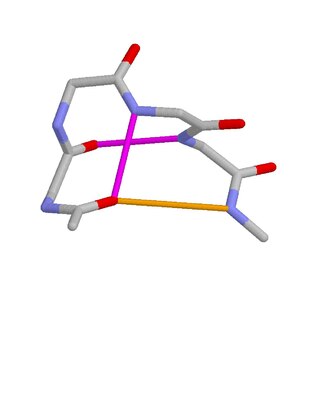
Schellman loops are commonly occurring structural features of proteins and polypeptides. Each has six amino acid residues with two specific inter-mainchain hydrogen bonds and a characteristic main chain dihedral angle conformation. The CO group of residue i is hydrogen-bonded to the NH of residue i+5, and the CO group of residue i+1 is hydrogen-bonded to the NH of residue i+4. Residues i+1, i+2, and i+3 have negative φ (phi) angle values and the phi value of residue i+4 is positive. Schellman loops incorporate a three amino acid residue RL nest, in which three mainchain NH groups form a concavity for hydrogen bonding to carbonyl oxygens. About 2.5% of amino acids in proteins belong to Schellman loops. Two websites are available for examining small motifs in proteins, Motivated Proteins: ; or PDBeMotif:.
The Asx turn is a structural feature in proteins and polypeptides. It consists of three amino acid residues in which residue i is an aspartate (Asp) or asparagine (Asn) that forms a hydrogen bond from its sidechain CO group to the mainchain NH group of residue i+2. About 14% of Asx residues present in proteins belong to Asx turns.















