The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand-helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located three or four residues earlier along the protein sequence.

The collagen triple helix or type-2 helix is the primary secondary structure of various types of fibrous collagen, including type I collagen. It consists of a triple helix made of the repetitious amino acid sequence glycine-X-Y, where X and Y are frequently proline or hydroxyproline. Collagen folded into a triple helix is known as tropocollagen. Collagen triple helices are often bundled into fibrils which themselves form larger fibres, as in tendon.

Protein secondary structure is the three dimensional form of local segments of proteins. The two most common secondary structural elements are alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein folds into its three dimensional tertiary structure.

Cyanobacteria, also known as Cyanophyta, are a phylum of prokaryotes consisting of both free-living photosynthetic bacteria and the endosymbiotic plastids that are present in the Archaeplastida, the autotrophic eukaryotes that include the red and green algae and land plants. They commonly obtain their energy through oxygenic photosynthesis, which produces the oxygen gas in the atmosphere of Earth. The name cyanobacteria comes from their color, giving them their other name, "blue-green algae", though some modern botanists restrict the term algae to eukaryotes. They appear to have originated in freshwater or a terrestrial environment.

A zinc finger is a small protein structural motif that is characterized by the coordination of one or more zinc ions (Zn2+) in order to stabilize the fold. Originally coined to describe the finger-like appearance of a hypothesized structure from Xenopus laevis transcription factor IIIA, the zinc finger name has now come to encompass a wide variety of differing protein structures. Xenopus laevis TFIIIA was originally demonstrated to contain zinc and require the metal for function in 1983, the first such reported zinc requirement for a gene regulatory protein. It often appears as a metal-binding domain in multi-domain proteins.
A coiled coil is a structural motif in proteins in which 2–7 alpha-helices are coiled together like the strands of a rope. Many coiled coil-type proteins are involved in important biological functions, such as the regulation of gene expression — e.g., transcription factors. Notable examples are the oncoproteins c-Fos and c-jun, as well as the muscle protein tropomyosin.
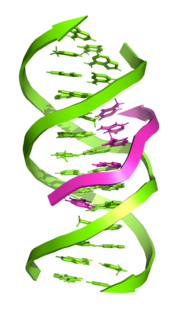
Triple-stranded DNA is a DNA structure in which three oligonucleotides wind around each other and form a triple helix. In triple-stranded DNA, the third strand binds to a B-form DNA double helix by forming Hoogsteen base pairs or reversed Hoogsteen hydrogen bonds.

A basic helix-loop-helix (bHLH) is a protein structural motif that characterizes one of the largest families of dimerizing transcription factors.

A leucine zipper is a common three-dimensional structural motif in proteins. They were first described by Landschulz and collaborators in 1988 when they found that an enhancer binding protein had a very characteristic 30-amino acid segment and the display of these amino acid sequences on an idealized alpha helix revealed a periodic repetition of leucine residues at every seventh position over a distance covering eight helical turns. The polypeptide segments containing these periodic arrays of leucine residues were proposed to exist in an alpha-helical conformation and the leucine side chains from one alpha helix interdigitate with those from the alpha helix of a second polypeptide, facilitating dimerization.
A DNA-binding domain (DBD) is an independently folded protein domain that contains at least one structural motif that recognizes double- or single-stranded DNA. A DBD can recognize a specific DNA sequence or have a general affinity to DNA. Some DNA-binding domains may also include nucleic acids in their folded structure.

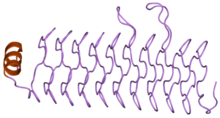
A beta helix is a tandem protein repeat structure formed by the association of parallel beta strands in a helical pattern with either two or three faces. The beta helix is a type of solenoid protein domain. The structure is stabilized by inter-strand hydrogen bonds, protein-protein interactions, and sometimes bound metal ions. Both left- and right-handed beta helices have been identified. Double stranded beta-helices are also very common features of proteins and are generally synonymous with jelly roll folds.
Forkhead box protein K2 is a protein that in humans is encoded by the FOXK2 gene.

The tetratricopeptide repeat (TPR) is a structural motif. It consists of a degenerate 34 amino acid tandem repeat identified in a wide variety of proteins. It is found in tandem arrays of 3–16 motifs, which form scaffolds to mediate protein–protein interactions and often the assembly of multiprotein complexes. These alpha-helix pair repeats usually fold together to produce a single, linear solenoid domain called a TPR domain. Proteins with such domains include the anaphase-promoting complex (APC) subunits cdc16, cdc23 and cdc27, the NADPH oxidase subunit p67-phox, hsp90-binding immunophilins, transcription factors, the protein kinase R (PKR), the major receptor for peroxisomal matrix protein import PEX5, protein arginine methyltransferase 9 (PRMT9), and mitochondrial import proteins.
RiAFP refers to an antifreeze protein (AFP) produced by the Rhagium inquisitor longhorned beetle. It is a type V antifreeze protein with a molecular weight of 12.8 kDa; this type of AFP is noted for its hyperactivity. R. inquisitor is a freeze-avoidant species, meaning that, due to its AFP, R. inquisitor prevents its body fluids from freezing altogether. This contrasts with freeze-tolerant species, whose AFPs simply depress levels of ice crystal formation in low temperatures. Whereas most insect antifreeze proteins contain cysteines at least every sixth residue, as well as varying numbers of 12- or 13-mer repeats of 8.3-12.5kDa, RiAFP is notable for containing only one disulfide bridge. This property of RiAFP makes it particularly attractive for recombinant expression and biotechnological applications.
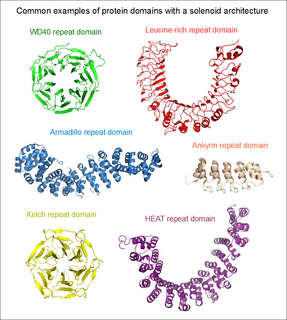
Solenoid protein domains are a highly modular type of protein domain. They consist of a chain of nearly identical folds, often simply called tandem repeats. They are extremely common among all types of proteins, though exact figures are unknown.
The term N cap describes an amino acid in a particular position within a protein or polypeptide. The N cap residue of an alpha helix is the first amino acid residue at the N terminus of the helix. More precisely, it is defined as the first residue (i) whose CO group is hydrogen-bonded to the NH group of residue i+4. Because of this it is sometimes also described as the residue prior to the helix.
Amide Rings are small motifs in proteins and polypeptides. They consist of 9-atom or 11-atom rings formed by two CO...HN hydrogen bonds between a side chain amide group and the main chain atoms of a short polypeptide. They are observed with glutamine or asparagine side chains within proteins and polypeptides. Structurally similar rings occur in the binding of purine, pyrimidine and nicotinamide bases to the main chain atoms of proteins. About 4% of asparagines and glutamines form amide rings; in databases of protein domain structures, one is present, on average, every other protein.
Cyanothece is a genus of unicellular, diazotrophic, oxygenic photosynthesizing cyanobacteria.

An array of protein tandem repeats is defined as several adjacent copies having the same or similar sequence motifs. These periodic sequences are generated by internal duplications in both coding and non-coding genomic sequences. Repetitive units of protein tandem repeats are considerably diverse, ranging from the repetition of a single amino acid to domains of 100 or more residues.

A toroid repeat is a protein fold composed of repeating subunits, arranged in circular fashion to form a closed structure.