
Crystallography is the experimental science of determining the arrangement of atoms in crystalline solids. The word "crystallography" is derived from the Greek words crystallon "cold drop, frozen drop", with its meaning extending to all solids with some degree of transparency, and graphein "to write". In July 2012, the United Nations recognised the importance of the science of crystallography by proclaiming that 2014 would be the International Year of Crystallography.

X-ray crystallography is the experimental science determining the atomic and molecular structure of a crystal, in which the crystalline structure causes a beam of incident X-rays to diffract into many specific directions. By measuring the angles and intensities of these diffracted beams, a crystallographer can produce a three-dimensional picture of the density of electrons within the crystal. From this electron density, the mean positions of the atoms in the crystal can be determined, as well as their chemical bonds, their crystallographic disorder, and various other information.
The Protein Data Bank (PDB) is a database for the three-dimensional structural data of large biological molecules, such as proteins and nucleic acids. The data, typically obtained by X-ray crystallography, NMR spectroscopy, or, increasingly, cryo-electron microscopy, and submitted by biologists and biochemists from around the world, are freely accessible on the Internet via the websites of its member organisations. The PDB is overseen by an organization called the Worldwide Protein Data Bank, wwPDB.
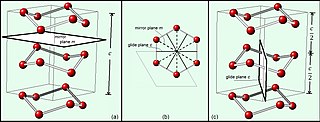
In mathematics, physics and chemistry, a space group is the symmetry group of an object in space, usually in three dimensions. The elements of a space group are the rigid transformations of an object that leave it unchanged. In three dimensions, space groups are classified into 219 distinct types, or 230 types if chiral copies are considered distinct. Space groups are discrete cocompact groups of isometries of an oriented Euclidean space in any number of dimensions. In dimensions other than 3, they are sometimes called Bieberbach groups.

Structural bioinformatics is the branch of bioinformatics that is related to the analysis and prediction of the three-dimensional structure of biological macromolecules such as proteins, RNA, and DNA. It deals with generalizations about macromolecular 3D structures such as comparisons of overall folds and local motifs, principles of molecular folding, evolution, binding interactions, and structure/function relationships, working both from experimentally solved structures and from computational models. The term structural has the same meaning as in structural biology, and structural bioinformatics can be seen as a part of computational structural biology. The main objective of structural bioinformatics is the creation of new methods of analysing and manipulating biological macromolecular data in order to solve problems in biology and generate new knowledge.

The International Centre for Diffraction Data (ICDD) maintains a database of powder diffraction patterns, the Powder Diffraction File (PDF), including the d-spacings and relative intensities of observable diffraction peaks. Patterns may be experimentally determined, or computed based on crystal structure and Bragg's law. It is most often used to identify substances based on x-ray diffraction data, and is designed for use with a diffractometer. Release 2022 of the Powder Diffraction File™ (PDF®) contains 1,076,439 unique material data sets. Each data set contains diffraction, crystallographic and bibliographic data, as well as experimental, instrument and sampling conditions, and select physical properties in a common standardized format.
Electron crystallography is a method to determine the arrangement of atoms in solids using a transmission electron microscope (TEM).

Aminocaproic acid is a derivative and analogue of the amino acid lysine, which makes it an effective inhibitor for enzymes that bind that particular residue. Such enzymes include proteolytic enzymes like plasmin, the enzyme responsible for fibrinolysis. For this reason it is effective in treatment of certain bleeding disorders, and it is sold under the brand name Amicar. Aminocaproic acid is also an intermediate in the polymerization of Nylon-6, where it is formed by ring-opening hydrolysis of caprolactam. The crystal structure determination showed that the 6-aminohexanoic acid is present as a salt, at least in the solid state.
Olga Kennard, née Weisz is a British scientist specialising in crystallography, and founder of the Cambridge Crystallographic Data Centre.
The Protein Data Bank (pdb) file format is a textual file format describing the three-dimensional structures of molecules held in the Protein Data Bank. The pdb format accordingly provides for description and annotation of protein and nucleic acid structures including atomic coordinates, secondary structure assignments, as well as atomic connectivity. In addition experimental metadata are stored. PDB format is the legacy file format for the Protein Data Bank which now keeps data on biological macromolecules in the newer mmCIF file format.
Crystallographic Information File (CIF) is a standard text file format for representing crystallographic information, promulgated by the International Union of Crystallography (IUCr). CIF was developed by the IUCr Working Party on Crystallographic Information in an effort sponsored by the IUCr Commission on Crystallographic Data and the IUCr Commission on Journals. The file format was initially published by Hall, Allen, and Brown and has since been revised, most recently versions 1.1 and 2.0. Full specifications for the format are available at the IUCr website. Many computer programs for molecular viewing are compatible with this format, including Jmol.

The Cambridge Structural Database (CSD) is both a repository and a validated and curated resource for the three-dimensional structural data of molecules generally containing at least carbon and hydrogen, comprising a wide range of organic, metal-organic and organometallic molecules. The specific entries are complementary to the other crystallographic databases such as the Protein Data Bank (PDB), Inorganic Crystal Structure Database and International Centre for Diffraction Data. The data, typically obtained by X-ray crystallography and less frequently by electron diffraction or neutron diffraction, and submitted by crystallographers and chemists from around the world, are freely accessible on the Internet via the CSD's parent organization's website. The CSD is overseen by the not-for-profit incorporated company called the Cambridge Crystallographic Data Centre, CCDC.

The Cambridge Crystallographic Data Centre (CCDC) is a non-profit organisation based in Cambridge, England. Its primary activity is the compilation and maintenance of the Cambridge Structural Database, a database of small molecule crystal structures. They also perform analysis on the database for the benefit of the scientific community, and write and distribute computer software to allow others to do the same.
Acta Crystallographica is a series of peer-reviewed scientific journals, with articles centred on crystallography, published by the International Union of Crystallography (IUCr). Originally established in 1948 as a single journal called Acta Crystallographica, there are now six independent Acta Crystallographica titles:
In biology, a protein structure database is a database that is modeled around the various experimentally determined protein structures. The aim of most protein structure databases is to organize and annotate the protein structures, providing the biological community access to the experimental data in a useful way. Data included in protein structure databases often includes three-dimensional coordinates as well as experimental information, such as unit cell dimensions and angles for x-ray crystallography determined structures. Though most instances, in this case either proteins or a specific structure determinations of a protein, also contain sequence information and some databases even provide means for performing sequence based queries, the primary attribute of a structure database is structural information, whereas sequence databases focus on sequence information, and contain no structural information for the majority of entries. Protein structure databases are critical for many efforts in computational biology such as structure based drug design, both in developing the computational methods used and in providing a large experimental dataset used by some methods to provide insights about the function of a protein.
A crystallographic database is a database specifically designed to store information about the structure of molecules and crystals. Crystals are solids having, in all three dimensions of space, a regularly repeating arrangement of atoms, ions, or molecules. They are characterized by symmetry, morphology, and directionally dependent physical properties. A crystal structure describes the arrangement of atoms, ions, or molecules in a crystal.
Olex and Olex2 are versatile software for crystallographic research. Olex used to be a research project developed during PhD to implement topological analysis of polymeric chemical structures and still is widely used around the world. Olex2 is an open source project with the C++ code portable to Windows, Mac and Linux. Although the projects share the common name they are not related at the source code level.

Macromolecular structure validation is the process of evaluating reliability for 3-dimensional atomic models of large biological molecules such as proteins and nucleic acids. These models, which provide 3D coordinates for each atom in the molecule, come from structural biology experiments such as x-ray crystallography or nuclear magnetic resonance (NMR). The validation has three aspects: 1) checking on the validity of the thousands to millions of measurements in the experiment; 2) checking how consistent the atomic model is with those experimental data; and 3) checking consistency of the model with known physical and chemical properties.
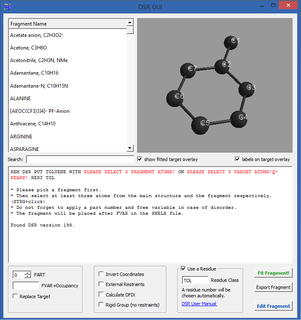
The Disordered Structure Refinement program (DSR), written by Daniel Kratzert, is designed to simplify the modeling of molecular disorder in crystal structures using SHELXL by George M. Sheldrick. It has a database of approximately 120 standard solvent molecules and molecular moieties. These can be inserted into the crystal structure with little effort, while at the same time chemically meaningful binding and angular restraints are set. DSR was developed because the previous description of disorder in crystal structures with SHELXL was very lengthy and error-prone. Instead of editing large text files manually and defining restraints manually, this process is automated with DSR.

CrystalExplorer or CE is a freeware designed to analysis the crystal structure with *.cif file format.











