Qt is cross-platform application development framework for creating graphical user interfaces as well as cross-platform applications that run on various software and hardware platforms such as Linux, Windows, macOS, Android or embedded systems with little or no change in the underlying codebase while still being a native application with native capabilities and speed.
Visual Molecular Dynamics (VMD) is a molecular modelling and visualization computer program. VMD is developed mainly as a tool to view and analyze the results of molecular dynamics simulations. It also includes tools for working with volumetric data, sequence data, and arbitrary graphics objects. Molecular scenes can be exported to external rendering tools such as POV-Ray, RenderMan, Tachyon, Virtual Reality Modeling Language (VRML), and many others. Users can run their own Tcl and Python scripts within VMD as it includes embedded Tcl and Python interpreters. VMD runs on Unix, Apple Mac macOS, and Microsoft Windows. VMD is available to non-commercial users under a distribution-specific license which permits both use of the program and modification of its source code, at no charge.

PyQt is a Python binding of the cross-platform GUI toolkit Qt, implemented as a Python plug-in. PyQt is free software developed by the British firm Riverbank Computing. It is available under similar terms to Qt versions older than 4.5; this means a variety of licenses including GNU General Public License (GPL) and commercial license, but not the GNU Lesser General Public License (LGPL). PyQt supports Microsoft Windows as well as various kinds of UNIX, including Linux and MacOS.

PyGTK is a set of Python wrappers for the GTK graphical user interface library. PyGTK is free software and licensed under the LGPL. It is analogous to PyQt/PySide and wxPython, the Python wrappers for Qt and wxWidgets, respectively. Its original author is GNOME developer James Henstridge. There are six people in the core development team, with various other people who have submitted patches and bug reports. PyGTK has been selected as the environment of choice for applications running on One Laptop Per Child systems.
RasMol is a computer program written for molecular graphics visualization intended and used mainly to depict and explore biological macromolecule structures, such as those found in the Protein Data Bank (PDB).
GRIB is a concise data format commonly used in meteorology to store historical and forecast weather data. It is standardized by the World Meteorological Organization's Commission for Basic Systems, known under number GRIB FM 92-IX, described in WMO Manual on Codes No.306. Currently there are three versions of GRIB. Version 0 was used to a limited extent by projects such as TOGA, and is no longer in operational use. The first edition is used operationally worldwide by most meteorological centers, for Numerical Weather Prediction output (NWP). A newer generation has been introduced, known as GRIB second edition, and data is slowly changing over to this format. Some of the second-generation GRIB is used for derived products distributed in the Eumetcast of Meteosat Second Generation. Another example is the NAM model.
Warren Lyford DeLano was a bioinformatician and an advocate for the increased adoption of open source practices in the sciences, and especially drug discovery, where advances which save time and resources can also potentially save lives.

BALL is a C++ class framework and set of algorithms and data structures for molecular modelling and computational structural bioinformatics, a Python interface to this library, and a graphical user interface to BALL, the molecule viewer BALLView.

ParaView is an open-source multiple-platform application for interactive, scientific visualization. It has a client–server architecture to facilitate remote visualization of datasets, and generates level of detail (LOD) models to maintain interactive frame rates for large datasets. It is an application built on top of the Visualization Toolkit (VTK) libraries. ParaView is an application designed for data parallelism on shared-memory or distributed-memory multicomputers and clusters. It can also be run as a single-computer application.

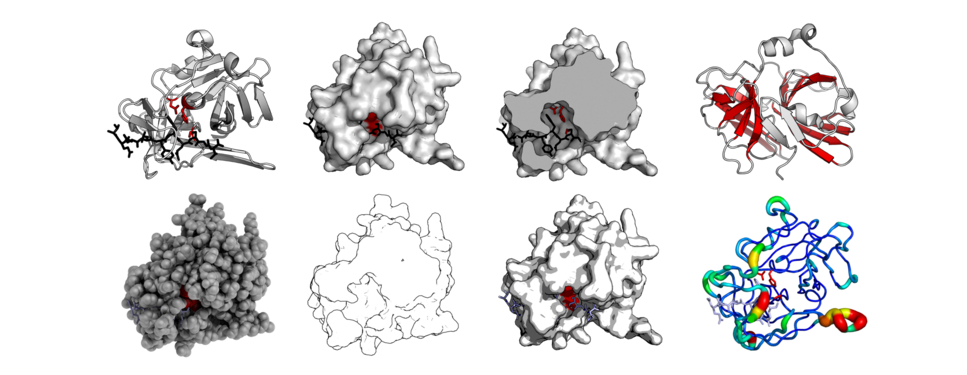
Ribbon diagrams, also known as Richardson diagrams, are 3D schematic representations of protein structure and are one of the most common methods of protein depiction used today. The ribbon depicts the general course and organisation of the protein backbone in 3D and serves as a visual framework for hanging details of the entire atomic structure, such as the balls for the oxygen atoms attached to myoglobin's active site in the adjacent figure. Ribbon diagrams are generated by interpolating a smooth curve through the polypeptide backbone. α-helices are shown as coiled ribbons or thick tubes, β-sheets as arrows, and non-repetitive coils or loops as lines or thin tubes. The direction of the polypeptide chain is shown locally by the arrows, and may be indicated overall by a colour ramp along the length of the ribbon.
Enthought, Inc. is a software company based in Austin, Texas, United States that develops scientific and analytic computing solutions using primarily the Python programming language. It is best known for the early development and maintenance of the SciPy library of mathematics, science, and engineering algorithms and for its Python for scientific computing distribution Enthought Canopy.

Avogadro is a molecule editor and visualizer designed for cross-platform use in computational chemistry, molecular modeling, bioinformatics, materials science, and related areas. It is extensible via a plugin architecture.

The Python Package Index, abbreviated as PyPI and also known as the Cheese Shop, is the official third-party software repository for Python. It is analogous to the CPAN repository for Perl and to the CRAN repository for R. PyPI is run by the Python Software Foundation, a charity. Some package managers, including pip, use PyPI as the default source for packages and their dependencies.
SIP is an open source software tool used to connect computer programs or libraries written in C or C++ with the scripting language Python. It is an alternative to SWIG.

PyCharm is an integrated development environment (IDE) used for programming in Python. It provides code analysis, a graphical debugger, an integrated unit tester, integration with version control systems, and supports web development with Django. PyCharm is developed by the Czech company JetBrains.

Spyder is an open-source cross-platform integrated development environment (IDE) for scientific programming in the Python language. Spyder integrates with a number of prominent packages in the scientific Python stack, including NumPy, SciPy, Matplotlib, pandas, IPython, SymPy and Cython, as well as other open-source software. It is released under the MIT license.

DelPhi is a scientific application which calculates electrostatic potentials in and around macromolecules and the corresponding electrostatic energies. It incorporates the effects of ionic strength mediated screening by evaluating the Poisson-Boltzmann equation at a finite number of points within a three-dimensional grid box. DelPhi is commonly used in protein science to visualize variations in electrostatics along a protein or other macromolecular surface and to calculate the electrostatic components of various energies.