The endoplasmic reticulum (ER) is, in essence, the transportation system of the eukaryotic cell, and has many other important functions such as protein folding. It is a type of organelle made up of two subunits – rough endoplasmic reticulum (RER), and smooth endoplasmic reticulum (SER). The endoplasmic reticulum is found in most eukaryotic cells and forms an interconnected network of flattened, membrane-enclosed sacs known as cisternae, and tubular structures in the SER. The membranes of the ER are continuous with the outer nuclear membrane. The endoplasmic reticulum is not found in red blood cells, or spermatozoa.
A congenital disorder of glycosylation is one of several rare inborn errors of metabolism in which glycosylation of a variety of tissue proteins and/or lipids is deficient or defective. Congenital disorders of glycosylation are sometimes known as CDG syndromes. They often cause serious, sometimes fatal, malfunction of several different organ systems in affected infants. The most common sub-type is PMM2-CDG where the genetic defect leads to the loss of phosphomannomutase 2 (PMM2), the enzyme responsible for the conversion of mannose-6-phosphate into mannose-1-phosphate.
A signal peptide is a short peptide present at the N-terminus of most newly synthesized proteins that are destined toward the secretory pathway. These proteins include those that reside either inside certain organelles, secreted from the cell, or inserted into most cellular membranes. Although most type I membrane-bound proteins have signal peptides, most type II and multi-spanning membrane-bound proteins are targeted to the secretory pathway by their first transmembrane domain, which biochemically resembles a signal sequence except that it is not cleaved. They are a kind of target peptide.
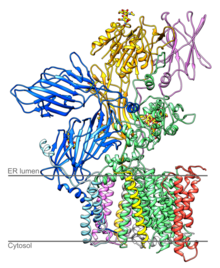
The translocon is a complex of proteins associated with the translocation of polypeptides across membranes. In eukaryotes the term translocon most commonly refers to the complex that transports nascent polypeptides with a targeting signal sequence into the interior space of the endoplasmic reticulum (ER) from the cytosol. This translocation process requires the protein to cross a hydrophobic lipid bilayer. The same complex is also used to integrate nascent proteins into the membrane itself. In prokaryotes, a similar protein complex transports polypeptides across the (inner) plasma membrane or integrates membrane proteins. In either case, the protein complex are formed from Sec proteins, with the heterotrimeric Sec61 being the channel. In prokaryotes, the homologous channel complex is known as SecYEG.
An oligosaccharide is a saccharide polymer containing a small number of monosaccharides. Oligosaccharides can have many functions including cell recognition and cell adhesion.
Dolichol refers to any of a group of long-chain mostly unsaturated organic compounds that are made up of varying numbers of isoprene units terminating in an α-saturated isoprenoid group, containing an alcohol functional group.

Ribophorins are dome shaped transmembrane glycoproteins which are located in the membrane of the rough endoplasmic reticulum, but are absent in the membrane of the smooth endoplasmic reticulum. There are two types of ribophorines: ribophorin I and II. These act in the protein complex oligosaccharyltransferase (OST) as two different subunits of the named complex. Ribophorin I and II are only present in eukaryote cells.

In enzymology, a dolichol kinase is an enzyme that catalyzes the chemical reaction

UDP-N-acetylglucosamine—dolichyl-phosphate N-acetylglucosaminephosphotransferase is an enzyme that in humans is encoded by the DPAGT1 gene.

Dolichyl-diphosphooligosaccharide—protein glycosyltransferase subunit 1 is an enzyme that in humans is encoded by the RPN1 gene.

Dolichyl-diphosphooligosaccharide—protein glycosyltransferase subunit 2, also called ribophorin ǁ is an enzyme that in humans is encoded by the RPN2 gene.

Nucleotide exchange factor SIL1 is a protein that in humans is encoded by the SIL1 gene.

Dolichol-phosphate mannosyltransferase is an enzyme that in humans is encoded by the DPM1 gene.
In cell biology, membrane bound polyribosomes are attached to a cell's endoplasmic reticulum. When certain proteins are synthesized by a ribosome they can become "membrane-bound". The newly produced polypeptide chains are inserted directly into the endoplasmic reticulum by the ribosome and are then transported to their destinations. Bound ribosomes usually produce proteins that are used within the cell membrane or are expelled from the cell via exocytosis.
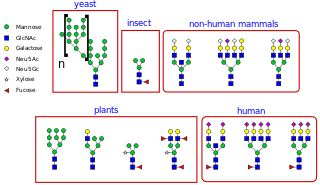
N-linked glycosylation, is the attachment of an oligosaccharide, a carbohydrate consisting of several sugar molecules, sometimes also referred to as glycan, to a nitrogen atom, in a process called N-glycosylation, studied in biochemistry. The resulting protein is called an N-linked glycan, or simply an N-glycan.

dolichyl-phosphate mannosyltransferase polypeptide 3, also known as DPM3, is a human gene.

In molecular biology, OST4 is a subunit of the oligosaccharyltransferase complex. OST4 is a very short, approximately 30 amino acids, protein found from fungi to vertebrates. It appears to be an integral membrane protein that mediates the en bloc transfer of a pre-assembled high-mannose oligosaccharide onto asparagine residues of nascent polypeptides as they enter the lumen of the rough endoplasmic reticulum.
Mannosyl-oligosaccharide glucosidase (MOGS) (EC 3.2.1.106, processing α-glucosidase I,Glc3Man9NAc2 oligosaccharide glucosidase, trimming glucosidase I, GCS1) is an enzyme with systematic name mannosyl-oligosaccharide glucohydrolase. MOGS is a transmembrane protein found in the membrane of the endoplasmic reticulum of eukaryotic cells. Biologically, it functions within the N-glycosylation pathway.

STT3A, catalytic subunit of the oligosaccharyltransferase complex is a protein that in humans is encoded by the STT3A gene.
William Joseph Lennarz was a biochemist at Stony Brook University. He was born in May 1934 in New York City. Before Lennarz began his tenure at Stony Brook, he studied chemistry and organic chemistry. After working as a postdoctoral researcher at Harvard, he developed an interest in biochemistry. He has focused the majority of his research on biochemical processes in cells.