A protein phosphatase is a phosphatase enzyme that removes a phosphate group from the phosphorylated amino acid residue of its substrate protein. Protein phosphorylation is one of the most common forms of reversible protein posttranslational modification (PTM), with up to 30% of all proteins being phosphorylated at any given time. Protein kinases (PKs) are the effectors of phosphorylation and catalyse the transfer of a γ-phosphate from ATP to specific amino acids on proteins. Several hundred PKs exist in mammals and are classified into distinct super-families. Proteins are phosphorylated predominantly on Ser, Thr and Tyr residues, which account for 79.3, 16.9 and 3.8% respectively of the phosphoproteome, at least in mammals. In contrast, protein phosphatases (PPs) are the primary effectors of dephosphorylation and can be grouped into three main classes based on sequence, structure and catalytic function. The largest class of PPs is the phosphoprotein phosphatase (PPP) family comprising PP1, PP2A, PP2B, PP4, PP5, PP6 and PP7, and the protein phosphatase Mg2+- or Mn2+-dependent (PPM) family, composed primarily of PP2C. The protein Tyr phosphatase (PTP) super-family forms the second group, and the aspartate-based protein phosphatases the third. The protein pseudophosphatases form part of the larger phosphatase family, and in most cases are thought to be catalytically inert, instead functioning as phosphate-binding proteins, integrators of signalling or subcellular traps. Examples of membrane-spanning protein phosphatases containing both active (phosphatase) and inactive (pseudophosphatase) domains linked in tandem are known, conceptually similar to the kinase and pseudokinase domain polypeptide structure of the JAK pseudokinases. A complete comparative analysis of human phosphatases and pseudophosphatases has been completed by Manning and colleagues, forming a companion piece to the ground-breaking analysis of the human kinome, which encodes the complete set of ~536 human protein kinases.

Receptor tyrosine kinases (RTKs) are the high-affinity cell surface receptors for many polypeptide growth factors, cytokines, and hormones. Of the 90 unique tyrosine kinase genes identified in the human genome, 58 encode receptor tyrosine kinase proteins. Receptor tyrosine kinases have been shown not only to be key regulators of normal cellular processes but also to have a critical role in the development and progression of many types of cancer. Mutations in receptor tyrosine kinases lead to activation of a series of signalling cascades which have numerous effects on protein expression. The receptors are generally activated by dimerization and substrate presentation. Receptor tyrosine kinases are part of the larger family of protein tyrosine kinases, encompassing the receptor tyrosine kinase proteins which contain a transmembrane domain, as well as the non-receptor tyrosine kinases which do not possess transmembrane domains.
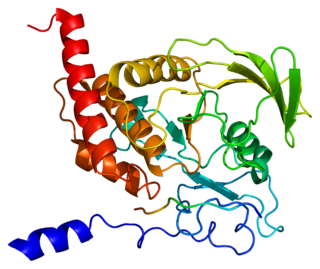
Tyrosine-protein phosphatase non-receptor type 6, also known as Src homology region 2 domain-containing phosphatase-1 (SHP-1), is an enzyme that in humans is encoded by the PTPN6 gene.
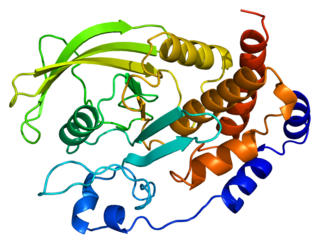
Tyrosine-protein phosphatase non-receptor type 1 also known as protein-tyrosine phosphatase 1B (PTP1B) is an enzyme that is the founding member of the protein tyrosine phosphatase (PTP) family. In humans it is encoded by the PTPN1 gene. PTP1B is a negative regulator of the insulin signaling pathway and is considered a promising potential therapeutic target, in particular for treatment of type 2 diabetes. It has also been implicated in the development of breast cancer and has been explored as a potential therapeutic target in that avenue as well.

Receptor-type tyrosine-protein phosphatase alpha is an enzyme that in humans is encoded by the PTPRA gene.

Receptor-type tyrosine-protein phosphatase F is an enzyme that, in humans, is encoded by the PTPRF gene.

Receptor-type tyrosine-protein phosphatase epsilon is an enzyme that in humans is encoded by the PTPRE gene.

Protein tyrosine phosphatase non-receptor type 7 is an enzyme that in humans is encoded by the PTPN7 gene.
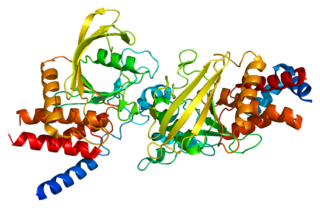
Receptor-type tyrosine-protein phosphatase beta or VE-PTP is an enzyme specifically expressed in endothelial cells that in humans is encoded by the PTPRB gene.

Receptor-type tyrosine-protein phosphatase mu is an enzyme that in humans is encoded by the PTPRM gene.

Receptor-type tyrosine-protein phosphatase PCP-2, is an enzyme that in humans is encoded by the PTPRU gene.
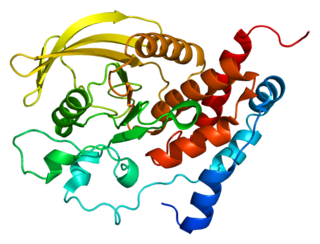
Protein tyrosine phosphatase receptor-type R is an enzyme that in humans is encoded by the PTPRR gene.

Liprin-alpha-1 is a protein that in humans is encoded by the PPFIA1 gene.

Receptor-type tyrosine-protein phosphatase kappa is an enzyme that in humans is encoded by the PTPRK gene. PTPRK is also known as PTPkappa and PTPκ.
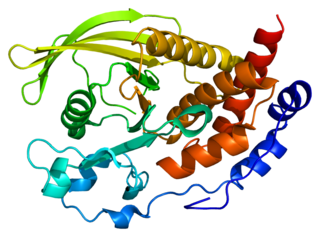
Protein tyrosine phosphatase non-receptor type 5 is an enzyme that in humans is encoded by the PTPN5 gene.

Serine/threonine-protein kinase LMTK2 also known as Lemur tyrosine kinase 2 (LMTK2) is an enzyme that in humans is encoded by the LMTK2 gene.
A non-receptor tyrosine kinase (nRTK) is a cytosolic enzyme that is responsible for catalysing the transfer of a phosphate group from a nucleoside triphosphate donor, such as ATP, to tyrosine residues in proteins. Non-receptor tyrosine kinases are a subgroup of protein family tyrosine kinases, enzymes that can transfer the phosphate group from ATP to a tyrosine residue of a protein (phosphorylation). These enzymes regulate many cellular functions by switching on or switching off other enzymes in a cell.

Autophosphorylation is a type of post-translational modification of proteins. It is generally defined as the phosphorylation of the kinase by itself. In eukaryotes, this process occurs by the addition of a phosphate group to serine, threonine or tyrosine residues within protein kinases, normally to regulate the catalytic activity. Autophosphorylation may occur when a kinases' own active site catalyzes the phosphorylation reaction, or when another kinase of the same type provides the active site that carries out the chemistry. The latter often occurs when kinase molecules dimerize. In general, the phosphate groups introduced are gamma phosphates from nucleoside triphosphates, most commonly ATP.

In molecular biology, YopH, N-terminal refers to an evolutionary conserved protein domain. This entry represents the N-terminal domain of YopH protein tyrosine phosphatase (PTP).

Tyrosine phosphorylation is the addition of a phosphate (PO43−) group to the amino acid tyrosine on a protein. It is one of the main types of protein phosphorylation. This transfer is made possible through enzymes called tyrosine kinases. Tyrosine phosphorylation is a key step in signal transduction and the regulation of enzymatic activity.






















