Beta-lactamases (β-lactamases) are enzymes produced by bacteria that provide multi-resistance to beta-lactam antibiotics such as penicillins, cephalosporins, cephamycins, monobactams and carbapenems (ertapenem), although carbapenems are relatively resistant to beta-lactamase. Beta-lactamase provides antibiotic resistance by breaking the antibiotics' structure. These antibiotics all have a common element in their molecular structure: a four-atom ring known as a beta-lactam (β-lactam) ring. Through hydrolysis, the enzyme lactamase breaks the β-lactam ring open, deactivating the molecule's antibacterial properties.

Enterobacteriaceae is a large family of Gram-negative bacteria. It includes over 30 genera and more than 100 species. Its classification above the level of family is still a subject of debate, but one classification places it in the order Enterobacterales of the class Gammaproteobacteria in the phylum Pseudomonadota. In 2016, the description and members of this family were emended based on comparative genomic analyses by Adeolu et al.

The cephalosporins are a class of β-lactam antibiotics originally derived from the fungus Acremonium, which was previously known as Cephalosporium.

A hospital-acquired infection, also known as a nosocomial infection, is an infection that is acquired in a hospital or other healthcare facility. To emphasize both hospital and nonhospital settings, it is sometimes instead called a healthcare-associated infection. Such an infection can be acquired in a hospital, nursing home, rehabilitation facility, outpatient clinic, diagnostic laboratory or other clinical settings. A number of dynamic processes can bring contamination into operating rooms and other areas within nosocomial settings. Infection is spread to the susceptible patient in the clinical setting by various means. Healthcare staff also spread infection, in addition to contaminated equipment, bed linens, or air droplets. The infection can originate from the outside environment, another infected patient, staff that may be infected, or in some cases, the source of the infection cannot be determined. In some cases the microorganism originates from the patient's own skin microbiota, becoming opportunistic after surgery or other procedures that compromise the protective skin barrier. Though the patient may have contracted the infection from their own skin, the infection is still considered nosocomial since it develops in the health care setting. Nosocomial infection tends to lack evidence that it was present when the patient entered the healthcare setting, thus meaning it was acquired post-admission.

Klebsiella is a genus of Gram-negative, oxidase-negative, rod-shaped bacteria with a prominent polysaccharide-based capsule.

Carbapenems are a class of very effective antibiotic agents most commonly used for treatment of severe bacterial infections. This class of antibiotics is usually reserved for known or suspected multidrug-resistant (MDR) bacterial infections. Similar to penicillins and cephalosporins, carbapenems are members of the beta-lactam antibiotics drug class, which kill bacteria by binding to penicillin-binding proteins, thus inhibiting bacterial cell wall synthesis. However, these agents individually exhibit a broader spectrum of activity compared to most cephalosporins and penicillins. Furthermore, carbapenems are typically unaffected by emerging antibiotic resistance, even to other beta-lactams.
Ampicillin/sulbactam is a fixed-dose combination medication of the common penicillin-derived antibiotic ampicillin and sulbactam, an inhibitor of bacterial beta-lactamase. Two different forms of the drug exist. The first, developed in 1987 and marketed in the United States under the brand name Unasyn, generic only outside the United States, is an intravenous antibiotic. The second, an oral form called sultamicillin, is marketed under the brand name Ampictam outside the United States, and generic only in the United States. Ampicillin/sulbactam is used to treat infections caused by bacteria resistant to beta-lactam antibiotics. Sulbactam blocks the enzyme which breaks down ampicillin and thereby allows ampicillin to attack and kill the bacteria.

Temocillin is a β-lactamase-resistant penicillin introduced by Beecham, marketed by Eumedica Pharmaceuticals as Negaban. It is used primarily for the treatment of multiple drug-resistant, Gram-negative bacteria.
It is a 6-methoxy penicillin; it is also a carboxypenicillin.

Klebsiella oxytoca is a Gram-negative, rod-shaped bacterium that is closely related to K. pneumoniae, from which it is distinguished by being indole-positive; it also has slightly different growth characteristics in that it is able to grow on melezitose, but not 3-hydroxybutyrate. It was first described in 1886 when it was isolated from sour milk and named Bacillus oxytocus perniciosus.

Acinetobacter baumannii is a typically short, almost round, rod-shaped (coccobacillus) Gram-negative bacterium. It is named after the bacteriologist Paul Baumann. It can be an opportunistic pathogen in humans, affecting people with compromised immune systems, and is becoming increasingly important as a hospital-derived (nosocomial) infection. While other species of the genus Acinetobacter are often found in soil samples, it is almost exclusively isolated from hospital environments. Although occasionally it has been found in environmental soil and water samples, its natural habitat is still not known.

Cefoxitin is a second-generation cephamycin antibiotic developed by Merck & Co., Inc. from Cephamycin C in the year following its discovery, 1972. It was synthesized in order to create an antibiotic with a broader spectrum. It is often grouped with the second-generation cephalosporins. Cefoxitin requires a prescription and as of 2010 is sold under the brand name Mefoxin by Bioniche Pharma, LLC. The generic version of cefoxitin is known as cefoxitin sodium.

Beta-lactamases are a family of enzymes involved in bacterial resistance to beta-lactam antibiotics. In bacterial resistance to beta-lactam antibiotics, the bacteria have beta-lactamase which degrade the beta-lactam rings, rendering the antibiotic ineffective. However, with beta-lactamase inhibitors, these enzymes on the bacteria are inhibited, thus allowing the antibiotic to take effect. Strategies for combating this form of resistance have included the development of new beta-lactam antibiotics that are more resistant to cleavage and the development of the class of enzyme inhibitors called beta-lactamase inhibitors. Although β-lactamase inhibitors have little antibiotic activity of their own, they prevent bacterial degradation of beta-lactam antibiotics and thus extend the range of bacteria the drugs are effective against.

NDM-1 is an enzyme that makes bacteria resistant to a broad range of beta-lactam antibiotics. These include the antibiotics of the carbapenem family, which are a mainstay for the treatment of antibiotic-resistant bacterial infections. The gene for NDM-1 is one member of a large gene family that encodes beta-lactamase enzymes called carbapenemases. Bacteria that produce carbapenemases are often referred to in the news media as "superbugs" because infections caused by them are difficult to treat. Such bacteria are usually sensitive only to polymyxins and tigecycline.
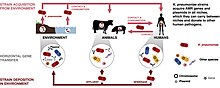
Plasmid-mediated resistance is the transfer of antibiotic resistance genes which are carried on plasmids. Plasmids possess mechanisms that ensure their independent replication as well as those that regulate their replication number and guarantee stable inheritance during cell division. By the conjugation process, they can stimulate lateral transfer between bacteria from various genera and kingdoms. Numerous plasmids contain addiction-inducing systems that are typically based on toxin-antitoxin factors and capable of killing daughter cells that don't inherit the plasmid during cell division. Plasmids often carry multiple antibiotic resistance genes, contributing to the spread of multidrug-resistance (MDR). Antibiotic resistance mediated by MDR plasmids severely limits the treatment options for the infections caused by Gram-negative bacteria, especially family Enterobacteriaceae. The global spread of MDR plasmids has been enhanced by selective pressure from antimicrobial medications used in medical facilities and when raising animals for food.
Multidrug resistant Gram-negative bacteria are a type of Gram-negative bacteria with resistance to multiple antibiotics. They can cause bacteria infections that pose a serious and rapidly emerging threat for hospitalized patients and especially patients in intensive care units. Infections caused by MDR strains are correlated with increased morbidity, mortality, and prolonged hospitalization. Thus, not only do these bacteria pose a threat to global public health, but also create a significant burden to healthcare systems.
Carbapenem-resistant Enterobacteriaceae (CRE) or carbapenemase-producing Enterobacteriaceae (CPE) are Gram-negative bacteria that are resistant to the carbapenem class of antibiotics, considered the drugs of last resort for such infections. They are resistant because they produce an enzyme called a carbapenemase that disables the drug molecule. The resistance can vary from moderate to severe. Enterobacteriaceae are common commensals and infectious agents. Experts fear CRE as the new "superbug". The bacteria can kill up to half of patients who get bloodstream infections. Tom Frieden, former head of the Centers for Disease Control and Prevention has referred to CRE as "nightmare bacteria". Examples of enzymes found in certain types of CRE are KPC and NDM. KPC and NDM are enzymes that break down carbapenems and make them ineffective. Both of these enzymes, as well as the enzyme VIM have also been reported in Pseudomonas.

Ceftazidime/avibactam, sold under the brand name Avycaz among others, is a fixed-dose combination medication composed of ceftazidime, a cephalosporin antibiotic, and avibactam, a β-lactamase inhibitor. It is used to treat complicated intra-abdominal infections, urinary tract infections, and pneumonia. It is only recommended when other options are not appropriate. It is given by infusion into a vein.
ESKAPE is an acronym comprising the scientific names of six highly virulent and antibiotic resistant bacterial pathogens including: Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp. The acronym is sometimes extended to ESKAPEE to include Escherichia coli. This group of Gram-positive and Gram-negative bacteria can evade or 'escape' commonly used antibiotics due to their increasing multi-drug resistance (MDR). As a result, throughout the world, they are the major cause of life-threatening nosocomial or hospital-acquired infections in immunocompromised and critically ill patients who are most at risk. P. aeruginosa and S. aureus are some of the most ubiquitous pathogens in biofilms found in healthcare. P. aeruginosa is a Gram-negative, rod-shaped bacterium, commonly found in the gut flora, soil, and water that can be spread directly or indirectly to patients in healthcare settings. The pathogen can also be spread in other locations through contamination, including surfaces, equipment, and hands. The opportunistic pathogen can cause hospitalized patients to have infections in the lungs, blood, urinary tract, and in other body regions after surgery. S. aureus is a Gram-positive, cocci-shaped bacterium, residing in the environment and on the skin and nose of many healthy individuals. The bacterium can cause skin and bone infections, pneumonia, and other types of potentially serious infections if it enters the body. S. aureus has also gained resistance to many antibiotic treatments, making healing difficult. Because of natural and unnatural selective pressures and factors, antibiotic resistance in bacteria usually emerges through genetic mutation or acquires antibiotic-resistant genes (ARGs) through horizontal gene transfer - a genetic exchange process by which antibiotic resistance can spread.
Cefiderocol, sold under the brand name Fetroja among others, is an antibiotic used to treat complicated urinary tract infections when no other options are available. It is indicated for the treatment of multi-drug-resistant Gram-negative bacteria including Pseudomonas aeruginosa. It is given by injection into a vein.