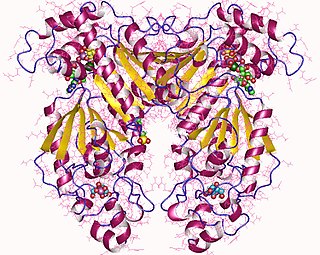
Phosphofructokinase-2 (6-phosphofructo-2-kinase, PFK-2) or fructose bisphosphatase-2 (FBPase-2), is an enzyme indirectly responsible for regulating the rates of glycolysis and gluconeogenesis in cells. It catalyzes formation and degradation of a significant allosteric regulator, fructose-2,6-bisphosphate (Fru-2,6-P2) from substrate fructose-6-phosphate. Fru-2,6-P2 contributes to the rate-determining step of glycolysis as it activates enzyme phosphofructokinase 1 in the glycolysis pathway, and inhibits fructose-1,6-bisphosphatase 1 in gluconeogenesis. Since Fru-2,6-P2 differentially regulates glycolysis and gluconeogenesis, it can act as a key signal to switch between the opposing pathways. Because PFK-2 produces Fru-2,6-P2 in response to hormonal signaling, metabolism can be more sensitively and efficiently controlled to align with the organism's glycolytic needs. This enzyme participates in fructose and mannose metabolism. The enzyme is important in the regulation of hepatic carbohydrate metabolism and is found in greatest quantities in the liver, kidney and heart. In mammals, several genes often encode different isoforms, each of which differs in its tissue distribution and enzymatic activity. The family described here bears a resemblance to the ATP-driven phospho-fructokinases, however, they share little sequence similarity, although a few residues seem key to their interaction with fructose 6-phosphate.
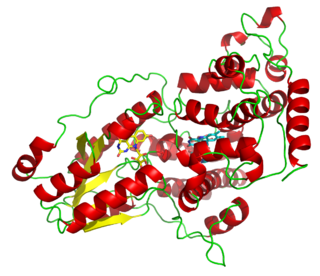
Photolyases are DNA repair enzymes that repair damage caused by exposure to ultraviolet light. These enzymes require visible light both for their own activation and for the actual DNA repair. The DNA repair mechanism involving photolyases is called photoreactivation. They mainly convert pyrimidine dimers into a normal pair of pyrimidine bases.
Paenarthrobacter ureafaciens KI72, popularly known as nylon-eating bacteria, is a strain of Paenarthrobacter ureafaciens that can digest certain by-products of nylon 6 manufacture. It uses a set of enzymes to digest nylon, popularly known as nylonase.

Histidine ammonia-lyase is an enzyme that in humans is encoded by the HAL gene. It converts histidine into ammonia and urocanic acid. Its systematic name is L-histidine ammonia-lyase (urocanate-forming).

The enzyme phenylalanine ammonia lyase (EC 4.3.1.24) catalyzes the conversion of L-phenylalanine to ammonia and trans-cinnamic acid.:
The enzyme S-(hydroxymethyl)glutathione synthase catalyzes the reaction
In enzymology, an aliphatic aldoxime dehydratase (EC 4.99.1.5) is an enzyme that catalyzes the chemical reaction
In enzymology, a phenylacetaldoxime dehydratase (EC 4.99.1.7) is an enzyme that catalyzes the chemical reaction

Isocitrate lyase, or ICL, is an enzyme in the glyoxylate cycle that catalyzes the cleavage of isocitrate to succinate and glyoxylate. Together with malate synthase, it bypasses the two decarboxylation steps of the tricarboxylic acid cycle and is used by bacteria, fungi, and plants.
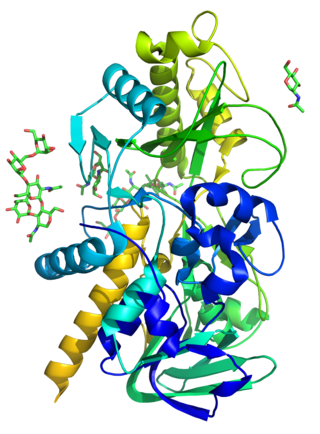
The enzyme (R)-mandelonitrile lyase (EC 4.1.2.10, (R)-HNL, (R)-oxynitrilase, (R)-hydroxynitrile lyase) catalyzes the chemical reaction
The enzyme chondroitin AC lyase catalyzes the chemical reaction
The enzyme chondroitin B lyase catalyzes the following process:
The enzyme levan fructotransferase (DFA-IV-forming) catalyzes the following process:
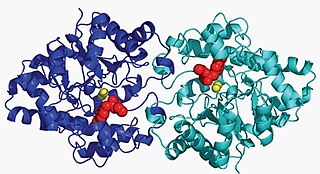
Aryldialkylphosphatase is a metalloenzyme that hydrolyzes the triester linkage found in organophosphate insecticides:
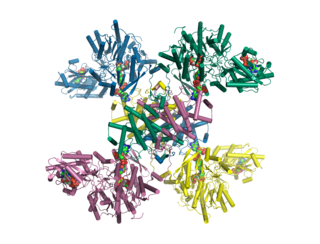
ATP citrate synthase (also ATP citrate lyase (ACLY)) is an enzyme that in animals represents an important step in fatty acid biosynthesis. By converting citrate to acetyl-CoA, the enzyme links carbohydrate metabolism, which yields citrate as an intermediate, with fatty acid biosynthesis, which consumes acetyl-CoA. In plants, ATP citrate lyase generates cytosolic acetyl-CoA precursors of thousands of specialized metabolites, including waxes, sterols, and polyketides.
Radical SAM is a designation for a superfamily of enzymes that use a [4Fe-4S]+ cluster to reductively cleave S-adenosyl-L-methionine (SAM) to generate a radical, usually a 5′-deoxyadenosyl radical (5'-dAdo), as a critical intermediate. These enzymes utilize this radical intermediate to perform diverse transformations, often to functionalize unactivated C-H bonds. Radical SAM enzymes are involved in cofactor biosynthesis, enzyme activation, peptide modification, post-transcriptional and post-translational modifications, metalloprotein cluster formation, tRNA modification, lipid metabolism, biosynthesis of antibiotics and natural products etc. The vast majority of known radical SAM enzymes belong to the radical SAM superfamily, and have a cysteine-rich motif that matches or resembles CxxxCxxC. rSAMs comprise the largest superfamily of metal-containing enzymes.
(S)-hydroxynitrile lyase (EC 4.1.2.47, (S)-cyanohydrin producing hydroxynitrile lyase, (S)-oxynitrilase, (S)-HbHNL, (S)-MeHNL, hydroxynitrile lyase, oxynitrilase, HbHNL, MeHNL, (S)-selective hydroxynitrile lyase, (S)-cyanohydrin carbonyl-lyase (cyanide forming), hydroxynitrilase) is an enzyme with systematic name (S)-cyanohydrin lyase (cyanide forming). This enzyme catalyses the interconversion between cyanohydrins and the carbonyl compounds derived from the cyanohydrin with free cyanide, as in the following two chemical reactions:
Aldos-2-ulose dehydratase (EC 4.2.1.110, pyranosone dehydratase, AUDH, 1,5-anhydro-D-fructose dehydratase (microthecin-forming)) is an enzyme with systematic name 1,5-anhydro-D-fructose hydro-lyase (microthecin-forming). This enzyme catalyses the following chemical reaction
UDP-N-acetylglucosamine 4,6-dehydratase (configuration-inverting) (EC 4.2.1.115, FlaA1, UDP-N-acetylglucosamine 5-inverting 4,6-dehydratase, PseB, UDP-N-acetylglucosamine hydro-lyase (inverting, UDP-2-acetamido-2,6-dideoxy-β-L)arabino-hex-4-ulose-forming)) is an enzyme with systematic name UDP-N-acetyl-α-D-glucosamine hydro-lyase (inverting; UDP-2-acetamido-2,6-dideoxy-β-L-arabino-hex-4-ulose-forming). This enzyme catalyses the following chemical reaction

Hydroperoxide lyases are enzymes that catalyze the cleavage of C-C bonds in the hydroperoxides of fatty acids. They belong to the cytochrome P450 enzyme family.









