
Protein biosynthesis is a core biological process, occurring inside cells, balancing the loss of cellular proteins through the production of new proteins. Proteins perform a number of critical functions as enzymes, structural proteins or hormones. Protein synthesis is a very similar process for both prokaryotes and eukaryotes but there are some distinct differences.

RNA editing is a molecular process through which some cells can make discrete changes to specific nucleotide sequences within an RNA molecule after it has been generated by RNA polymerase. It occurs in all living organisms and is one of the most evolutionarily conserved properties of RNAs. RNA editing may include the insertion, deletion, and base substitution of nucleotides within the RNA molecule. RNA editing is relatively rare, with common forms of RNA processing not usually considered as editing. It can affect the activity, localization as well as stability of RNAs, and has been linked with human diseases.
RNA-binding proteins are proteins that bind to the double or single stranded RNA in cells and participate in forming ribonucleoprotein complexes. RBPs contain various structural motifs, such as RNA recognition motif (RRM), dsRNA binding domain, zinc finger and others. They are cytoplasmic and nuclear proteins. However, since most mature RNA is exported from the nucleus relatively quickly, most RBPs in the nucleus exist as complexes of protein and pre-mRNA called heterogeneous ribonucleoprotein particles (hnRNPs). RBPs have crucial roles in various cellular processes such as: cellular function, transport and localization. They especially play a major role in post-transcriptional control of RNAs, such as: splicing, polyadenylation, mRNA stabilization, mRNA localization and translation. Eukaryotic cells express diverse RBPs with unique RNA-binding activity and protein–protein interaction. According to the Eukaryotic RBP Database (EuRBPDB), there are 2961 genes encoding RBPs in humans. During evolution, the diversity of RBPs greatly increased with the increase in the number of introns. Diversity enabled eukaryotic cells to utilize RNA exons in various arrangements, giving rise to a unique RNP (ribonucleoprotein) for each RNA. Although RBPs have a crucial role in post-transcriptional regulation in gene expression, relatively few RBPs have been studied systematically.It has now become clear that RNA–RBP interactions play important roles in many biological processes among organisms.

Potassium voltage-gated channel subfamily A member 1 also known as Kv1.1 is a shaker related voltage-gated potassium channel that in humans is encoded by the KCNA1 gene. Isaacs syndrome is a result of an autoimmune reaction against the Kv1.1 ion channel.
Missense mRNA is a messenger RNA bearing one or more mutated codons that yield polypeptides with an amino acid sequence different from the wild-type or naturally occurring polypeptide. Missense mRNA molecules are created when template DNA strands or the mRNA strands themselves undergo a missense mutation in which a protein coding sequence is mutated and an altered amino acid sequence is coded for.

Glutamate receptor 3 is a protein that in humans is encoded by the GRIA3 gene.
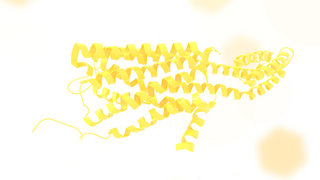
The 5-HT2C receptor is a subtype of 5-HT receptor that binds the endogenous neurotransmitter serotonin (5-hydroxytryptamine, 5-HT). It is a G protein-coupled receptor (GPCR) that is coupled to Gq/G11 and mediates excitatory neurotransmission. HTR2C denotes the human gene encoding for the receptor, that in humans is located at the X chromosome. As males have one copy of the gene and in females one of the two copies of the gene is repressed, polymorphisms at this receptor can affect the two sexes to differing extent.

Filamin A, alpha (FLNA) is a protein that in humans is encoded by the FLNA gene.

The double-stranded RNA-specific adenosine deaminase enzyme family are encoded by the ADAR family genes. ADAR stands for adenosine deaminase acting on RNA. This article focuses on the ADAR proteins; This article details the evolutionary history, structure, function, mechanisms and importance of all proteins within this family.

Glutamate ionotropic receptor AMPA type subunit 2 is a protein that in humans is encoded by the GRIA2 gene and it is a subunit found in the AMPA receptors.

Glutamate ionotropic receptor kainate type subunit 2, also known as ionotropic glutamate receptor 6 or GluR6, is a protein that in humans is encoded by the GRIK2 gene.

Insulin-like growth factor-binding protein 7 is a protein that in humans is encoded by the IGFBP7 gene. The major function of the protein is the regulation of availability of insulin-like growth factors (IGFs) in tissue as well as in modulating IGF binding to its receptors. IGFBP7 binds to IGF with low affinity compared to IGFBPs 1-6. It also stimulates cell adhesion. The protein is implicated in some cancers.

Double-stranded RNA-specific editase 1 is an enzyme that in humans is encoded by the ADARB1 gene. The enzyme is a member of ADAR family.

Glutamate receptor, ionotropic, kainate 1, also known as GRIK1, is a protein that in humans is encoded by the GRIK1 gene.

Cytoplasmic FMR1-interacting protein 2 is a protein that in humans is encoded by the CYFIP2 gene. Cytoplasmic FMR1 interacting protein is a 1253 amino acid long protein and is highly conserved sharing 99% sequence identity to the mouse protein. It is expressed mainly in brain tissues, white blood cells and the kidney.

Glutamate receptor 4 is a protein that in humans is encoded by the GRIA4 gene.

Gamma-aminobutyric acid receptor subunit alpha-3 is a protein that in humans is encoded by the GABRA3 gene.

Bladder cancer-associated protein is a protein that in humans is encoded by the BLCAP gene.
Within the science of molecular biology and cell biology, for human genetics, the GRIA2 gene is located on chromosome 4q32-q33. The gene product is the ionotropic AMPA glutamate receptor 2. The protein belongs to a family of ligand-activated glutamate receptors that are sensitive to alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate (AMPA). Glutamate receptors function as the main excitatory neurotransmitter at many synapses in the central nervous system. L-glutamate, an excitatory neurotransmitter, binds to the Gria2 resulting in a conformational change. This leads to the opening of the channel converting the chemical signal to an electrical impulse. AMPA receptors (AMPAR) are composed of four subunits, designated as GluR1 (GRIA1), GluR2 (GRIA2), GluR3 (GRIA3), and GluR4(GRIA4) which combine to form tetramers. They are usually heterotrimeric but can be homodimeric. Each AMPAR has four sites to which an agonist can bind, one for each subunit.[5]

The complement component 1, q subcomponent-like 1 is encoded by a gene located at chromosome 17q21.31. It is a secreted protein and is 258 amino acids in length. The protein is widely expressed but its expression is highest in the brain and may also be involved in regulation of motor control. The pre-mRNA of this protein is subject to RNA editing.



















