
Enterobacteriaceae is a large family of Gram-negative bacteria. It includes over 30 genera and more than 100 species. Its classification above the level of family is still a subject of debate, but one classification places it in the order Enterobacterales of the class Gammaproteobacteria in the phylum Pseudomonadota. In 2016, the description and members of this family were emended based on comparative genomic analyses by Adeolu et al.

A microbiological culture, or microbial culture, is a method of multiplying microbial organisms by letting them reproduce in predetermined culture medium under controlled laboratory conditions. Microbial cultures are foundational and basic diagnostic methods used as research tools in molecular biology.

Proteus vulgaris is a rod-shaped, nitrate-reducing, indole-positive and catalase-positive, hydrogen sulfide-producing, Gram-negative bacterium that inhabits the intestinal tracts of humans and animals. It can be found in soil, water, and fecal matter. It is grouped with the Morganellaceae and is an opportunistic pathogen of humans. It is known to cause wound infections and other species of its genera are known to cause urinary tract infections.
Plesiomonas shigelloides is a species of bacteria and the only member of its genus. It is a Gram-negative, rod-shaped bacterium which has been isolated from freshwater, freshwater fish, shellfish, cattle, goats, swine, cats, dogs, monkeys, vultures, snakes, toads and humans. It is considered a fecal coliform. P. shigelloides is a global distributed species, found globally outside of the polar ice caps.

A blood culture is a medical laboratory test used to detect bacteria or fungi in a person's blood. Under normal conditions, the blood does not contain microorganisms: their presence can indicate a bloodstream infection such as bacteremia or fungemia, which in severe cases may result in sepsis. By culturing the blood, microbes can be identified and tested for resistance to antimicrobial drugs, which allows clinicians to provide an effective treatment.

Coliform bacteria are defined as either motile or non-motile Gram-negative non-spore forming bacilli that possess β-galactosidase to produce acids and gases under their optimal growth temperature of 35–37 °C. They can be aerobes or facultative aerobes, and are a commonly used indicator of low sanitary quality of foods, milk, and water. Coliforms can be found in the aquatic environment, in soil and on vegetation; they are universally present in large numbers in the feces of warm-blooded animals as they are known to inhabit the gastrointestinal system. While coliform bacteria are not normally the cause of serious illness, they are easy to culture, and their presence is used to infer that other pathogenic organisms of fecal origin may be present in a sample, or that said sample is not safe to consume. Such pathogens include disease-causing bacteria, viruses, or protozoa and many multicellular parasites. Every drinking water source must be tested for the presence of these total coliform bacteria.

Shigella dysenteriae is a species of the rod-shaped bacterial genus Shigella. Shigella species can cause shigellosis. Shigellae are Gram-negative, non-spore-forming, facultatively anaerobic, nonmotile bacteria. S. dysenteriae has the ability to invade and replicate in various species of epithelial cells and enterocytes.
Bacillary dysentery is a type of dysentery, and is a severe form of shigellosis. It is associated with species of bacteria from the family Enterobacteriaceae. The term is usually restricted to Shigella infections.
The oxidase test is used to determine whether an organism possesses the cytochrome c oxidase enzyme. The test is used as an aid for the differentiation of Neisseria, Moraxella, Campylobacter and Pasteurella species. It is also used to differentiate pseudomonads from related species.

Antibiotic sensitivity testing or antibiotic susceptibility testing is the measurement of the susceptibility of bacteria to antibiotics. It is used because bacteria may have resistance to some antibiotics. Sensitivity testing results can allow a clinician to change the choice of antibiotics from empiric therapy, which is when an antibiotic is selected based on clinical suspicion about the site of an infection and common causative bacteria, to directed therapy, in which the choice of antibiotic is based on knowledge of the organism and its sensitivities.
Microbial metabolism is the means by which a microbe obtains the energy and nutrients it needs to live and reproduce. Microbes use many different types of metabolic strategies and species can often be differentiated from each other based on metabolic characteristics. The specific metabolic properties of a microbe are the major factors in determining that microbe's ecological niche, and often allow for that microbe to be useful in industrial processes or responsible for biogeochemical cycles.
In microbiology, the minimum inhibitory concentration (MIC) is the lowest concentration of a chemical, usually a drug, which prevents visible in vitro growth of bacteria or fungi. MIC testing is performed in both diagnostic and drug discovery laboratories.
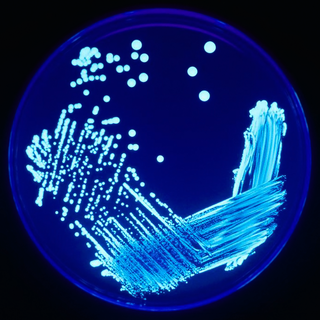
In microbiology, streaking is a technique used to isolate a pure strain from a single species of microorganism, often bacteria. Samples can then be taken from the resulting colonies and a microbiological culture can be grown on a new plate so that the organism can be identified, studied, or tested.

Etest is a way of determining antimicrobial sensitivity by placing a strip impregnated with antimicrobials onto an agar plate. A strain of bacterium or fungus will not grow near a concentration of antibiotic or antifungal if it is sensitive. For some microbial and antimicrobial combinations, the results can be used to determine a minimum inhibitory concentration (MIC). Etest is a proprietary system manufactured by bioMérieux. It is a laboratory test used in healthcare settings to help guide physicians by indicating what concentration of antimicrobial could successfully be used to treat patients' infections.

The IMViC tests are a group of individual tests used in microbiology lab testing to identify an organism in the coliform group. A coliform is a gram negative, aerobic, or facultative anaerobic rod, which produces gas from lactose within 48 hours. The presence of some coliforms indicate fecal contamination.
Edwardsiella tarda is a member of the family Hafniaceae. The bacterium is a facultatively anaerobic, small, motile, gram negative, straight rod with flagella. Infection causes Edwardsiella septicemia in channel catfish, eels, and flounder. Edwardsiella tarda is also found in largemouth bass and freshwater species such as rainbow trout. It is a zoonosis and can infect a variety of animals including fish, amphibians, reptiles, and mammals. Edwardsiella tarda has also been the cause of periodic infections for various animals within zoos. E. tarda has a worldwide distribution and can be found in pond water, mud, and the intestine of fish and other marine animals. It is spread by carrier animal feces.

Neisseria flavescens was first isolated from cerebrospinal fluid in the midst of an epidemic meningitis outbreak in Chicago. These gram-negative, aerobic bacteria reside in the mucosal membranes of the upper respiratory tract, functioning as commensals. However, this species can also play a pathogenic role in immunocompromised and diabetic individuals. In rare cases, it has been linked to meningitis, pneumonia, empyema, endocarditis, and sepsis.
In microbiology, the term isolation refers to the separation of a strain from a natural, mixed population of living microbes, as present in the environment, for example in water or soil, or from living beings with skin flora, oral flora or gut flora, in order to identify the microbe(s) of interest. Historically, the laboratory techniques of isolation first developed in the field of bacteriology and parasitology, before those in virology during the 20th century.
Diagnostic microbiology is the study of microbial identification. Since the discovery of the germ theory of disease, scientists have been finding ways to harvest specific organisms. Using methods such as differential media or genome sequencing, physicians and scientists can observe novel functions in organisms for more effective and accurate diagnosis of organisms. Methods used in diagnostic microbiology are often used to take advantage of a particular difference in organisms and attain information about what species it can be identified as, which is often through a reference of previous studies. New studies provide information that others can reference so that scientists can attain a basic understanding of the organism they are examining.

The Erwiniaceae are a family of Gram-negative bacteria which includes a number of plant pathogens and insect endosymbionts. This family is a member of the order Enterobacterales in the class Gammaproteobacteria of the phylum Pseudomonadota. The type genus of this family is Erwinia.












