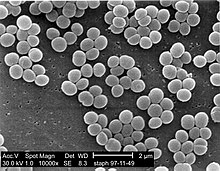
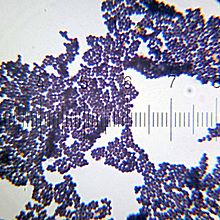
Staphylococcus aureus is a gram-positive spherically shaped bacterium, a member of the Bacillota, and is a usual member of the microbiota of the body, frequently found in the upper respiratory tract and on the skin. It is often positive for catalase and nitrate reduction and is a facultative anaerobe, meaning that it can grow without oxygen. Although S. aureus usually acts as a commensal of the human microbiota, it can also become an opportunistic pathogen, being a common cause of skin infections including abscesses, respiratory infections such as sinusitis, and food poisoning. Pathogenic strains often promote infections by producing virulence factors such as potent protein toxins, and the expression of a cell-surface protein that binds and inactivates antibodies. S. aureus is one of the leading pathogens for deaths associated with antimicrobial resistance and the emergence of antibiotic-resistant strains, such as methicillin-resistant S. aureus (MRSA). The bacterium is a worldwide problem in clinical medicine. Despite much research and development, no vaccine for S. aureus has been approved.

Coagulase is a protein enzyme produced by several microorganisms that enables the conversion of fibrinogen to fibrin. In the laboratory, it is used to distinguish between different types of Staphylococcus isolates. Importantly, S. aureus is generally coagulase-positive, meaning that a positive coagulase test would indicate the presence of S. aureus or any of the other 11 coagulase-positive Staphylococci. A negative coagulase test would instead show the presence of coagulase-negative organisms such as S. epidermidis or S. saprophyticus. However, it is now known that not all S. aureus are coagulase-positive. Whereas coagulase-positive staphylococci are usually pathogenic, coagulase-negative staphylococci are more often associated with opportunistic infection.
Staphylococcus lugdunensis is a coagulase-negative member of the genus Staphylococcus, consisting of Gram-positive bacteria with spherical cells that appear in clusters.

Staphylococcus haemolyticus is a member of the coagulase-negative staphylococci (CoNS). It is part of the skin flora of humans, and its largest populations are usually found at the axillae, perineum, and inguinal areas. S. haemolyticus also colonizes primates and domestic animals. It is a well-known opportunistic pathogen, and is the second-most frequently isolated CoNS. Infections can be localized or systemic, and are often associated with the insertion of medical devices. The highly antibiotic-resistant phenotype and ability to form biofilms make S. haemolyticus a difficult pathogen to treat. Its most closely related species is Staphylococcus borealis.
Staphylococcus saprophyticus is a Gram-positive coccus belonging to the genus Staphylococcus. S. saprophyticus is a common cause of community-acquired urinary tract infections.
Staphylococcus caprae is a Gram-positive, coccus bacteria and a member of the genus Staphylococcus. S. caprae is coagulase-negative. It was originally isolated from goats, but members of this species have also been isolated from human samples.
Staphylococcus hominis is a coagulase-negative member of the bacterial genus Staphylococcus, consisting of Gram-positive, spherical cells in clusters. It occurs very commonly as a harmless commensal on human and animal skin and is known for producing thioalcohol compounds that contribute to body odour. Like many other coagulase-negative staphylococci, S. hominis may occasionally cause infection in patients whose immune systems are compromised, for example by chemotherapy or predisposing illness.

Staphylococcus xylosus is a species of bacteria belonging to the genus Staphylococcus. It is a Gram-positive bacterium that forms clusters of cells. Like most staphylococcal species, it is coagulase-negative and exists as a commensal on the skin of humans and animals and in the environment.
Staphylococcus warneri is a member of the bacterial genus Staphylococcus, consisting of Gram-positive bacteria with spherical cells appearing in clusters. It is catalase-positive, oxidase-negative, and coagulase-negative, and is a common commensal organism found as part of the skin flora on humans and animals. Like other coagulase-negative staphylococci, S. warneri rarely causes disease, but may occasionally cause infection in patients whose immune system is compromised.

Staphylococcus epidermidis is a Gram-positive bacterium, and one of over 40 species belonging to the genus Staphylococcus. It is part of the normal human microbiota, typically the skin microbiota, and less commonly the mucosal microbiota and also found in marine sponges. It is a facultative anaerobic bacteria. Although S. epidermidis is not usually pathogenic, patients with compromised immune systems are at risk of developing infection. These infections are generally hospital-acquired. S. epidermidis is a particular concern for people with catheters or other surgical implants because it is known to form biofilms that grow on these devices. Being part of the normal skin microbiota, S. epidermidis is a frequent contaminant of specimens sent to the diagnostic laboratory.
A staphylococcal infection or staph infection is an infection caused by members of the Staphylococcus genus of bacteria.
mecA is a gene found in bacterial cells which allows them to be resistant to antibiotics such as methicillin, penicillin and other penicillin-like antibiotics.

Staphylococcus capitis is a coagulase-negative species (CoNS) of Staphylococcus. It is part of the normal flora of the skin of the human scalp, face, neck, scrotum, and ears and has been associated with prosthetic valve endocarditis, but is rarely associated with native valve infection.
Staphylococcus carnosus is a bacterium from the genus Staphylococcus that is both Gram-positive and coagulase-negative. It was originally identified in dry sausage and is an important starter culture for meat fermentation. Unlike other members of its genus, such as Staphylococcus aureus and Staphylococcus epidermidis, S. carnosus is nonpathogenic and safely used in the food industry.
Staphylococcus delphini is a Gram-positive, coagulase-positive member of the bacterial genus Staphylococcus consisting of single, paired, and clustered cocci. Strains of this species were originally isolated from aquarium-raised dolphins suffering from skin lesions.
Staphylococcus intermedius is a Gram-positive, catalase positive member of the bacterial genus Staphylococcus consisting of clustered cocci. Strains of this species were originally isolated from the anterior nares of pigeons, dogs, cats, mink, and horses. Many of the isolated strains show coagulase activity. Clinical tests for detection of methicillin-resistant S. aureus may produce false positives by detecting S. intermedius, as this species shares some phenotypic traits with methicillin-resistant S. aureus strains. It has been theorized that S. intermedius has previously been misidentified as S. aureus in human dog bite wound infections, which is why molecular technologies such as MALDI-TOF and PCR are preferred in modern veterinary clinical microbiology laboratories for their more accurate identifications over biochemical tests. S. intermedius is largely phenotypically indiscriminate from Staphylococcus pseudintermedius and Staphylococcus delphini, and therefore the three organisms are considered to be included in the more general 'Staphylococcus intermedius group'.
Staphylococcus schleiferi is a Gram-positive, cocci-shaped bacterium of the family Staphylococcaceae. It is facultatively anaerobic, coagulase-variable, and can be readily cultured on blood agar where the bacterium tends to form opaque, non-pigmented colonies and beta (β) hemolysis. There exists two subspecies under the species S. schleiferi: Staphylococcus schleiferi subsp. schleiferi and Staphylococcus schleiferi subsp. coagulans.
Staphylococcus pseudintermedius is a gram-positive spherically shaped bacterium of the genus Staphylococcus found worldwide. It is primarily a pathogen for domestic animals, but has been known to affect humans as well. S. pseudintermedius is an opportunistic pathogen that secretes immune-modulating virulence factors, has many adhesion factors, and the potential to create biofilms, all of which help to determine the pathogenicity of the bacterium. Diagnoses of S. pseudintermedius have traditionally been made using cytology, plating, and biochemical tests. More recently, molecular technologies like MALDI-TOF, DNA hybridization and PCR have become preferred over biochemical tests for their more rapid and accurate identifications. This includes the identification and diagnosis of antibiotic resistant strains.

Georg Peters was a German physician, microbiologist and university professor. From 1992 until his fatal mountain accident he headed the Institute of Medical Microbiology at the University of Münster. He was an internationally recognised expert in the field of staphylococci and the infectious diseases caused by them, to which he had devoted himself since the beginning of his scientific career.
Staphylococcus cornubiensis is a species of Gram-positive cocci in the Staphylococcus intermedius Group (SIG): a group of genetically and phenotypically similar bacterial species that were previously identified as S. intermedius. The bacterium was first isolated from a human skin infection in Cornwall, United Kingdom. However, its presence in other species and/or pathologies has yet to be discussed in the literature. Another SIG bacterium, S. pseudintermedius, has also been implicated in cutaneous infections in humans–as a result of zoonotic transmission from domestic animals. The other SIG species have been isolated from various wild and domestic animals; such as dogs, cats, horses, camels, and dolphins, among others.