
Staphylococcus aureus is a gram-positive spherically shaped bacterium, a member of the Bacillota, and is a usual member of the microbiota of the body, frequently found in the upper respiratory tract and on the skin. It is often positive for catalase and nitrate reduction and is a facultative anaerobe, meaning that it can grow without oxygen. Although S. aureus usually acts as a commensal of the human microbiota, it can also become an opportunistic pathogen, being a common cause of skin infections including abscesses, respiratory infections such as sinusitis, and food poisoning. Pathogenic strains often promote infections by producing virulence factors such as potent protein toxins, and the expression of a cell-surface protein that binds and inactivates antibodies. S. aureus is one of the leading pathogens for deaths associated with antimicrobial resistance and the emergence of antibiotic-resistant strains, such as methicillin-resistant S. aureus (MRSA). The bacterium is a worldwide problem in clinical medicine. Despite much research and development, no vaccine for S. aureus has been approved.
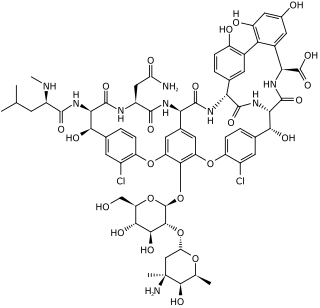
Vancomycin is a glycopeptide antibiotic medication used to treat a number of bacterial infections. It is used intravenously as a treatment for complicated skin infections, bloodstream infections, endocarditis, bone and joint infections, and meningitis caused by methicillin-resistant Staphylococcus aureus. Blood levels may be measured to determine the correct dose. Vancomycin is also taken orally as a treatment for severe Clostridium difficile colitis. When taken orally it is poorly absorbed.

Methicillin-resistant Staphylococcus aureus (MRSA) is a group of gram-positive bacteria that are genetically distinct from other strains of Staphylococcus aureus. MRSA is responsible for several difficult-to-treat infections in humans. It caused more than 100,000 deaths worldwide attributable to antimicrobial resistance in 2019.

Linezolid is an antibiotic used for the treatment of infections caused by Gram-positive bacteria that are resistant to other antibiotics. Linezolid is active against most Gram-positive bacteria that cause disease, including streptococci, vancomycin-resistant enterococci (VRE), and methicillin-resistant Staphylococcus aureus (MRSA). The main uses are infections of the skin and pneumonia although it may be used for a variety of other infections including drug-resistant tuberculosis. It is used either by injection into a vein or by mouth.

Methicillin (USAN), also known as meticillin (INN), is a narrow-spectrum β-lactam antibiotic of the penicillin class.

Glycopeptide antibiotics are a class of drugs of microbial origin that are composed of glycosylated cyclic or polycyclic nonribosomal peptides. Significant glycopeptide antibiotics include the anti-infective antibiotics vancomycin, teicoplanin, telavancin, ramoplanin and decaplanin, corbomycin, complestatin and the antitumor antibiotic bleomycin. Vancomycin is used if infection with methicillin-resistant Staphylococcus aureus (MRSA) is suspected.

Staphylococcus haemolyticus is a member of the coagulase-negative staphylococci (CoNS). It is part of the skin flora of humans, and its largest populations are usually found at the axillae, perineum, and inguinal areas. S. haemolyticus also colonizes primates and domestic animals. It is a well-known opportunistic pathogen, and is the second-most frequently isolated CoNS. Infections can be localized or systemic, and are often associated with the insertion of medical devices. The highly antibiotic-resistant phenotype and ability to form biofilms make S. haemolyticus a difficult pathogen to treat. Its most closely related species is Staphylococcus borealis.

Tigecycline, sold under the brand name Tygacil, is a tetracycline antibiotic medication for a number of bacterial infections. It is a glycylcycline class drug that is administered intravenously. It was developed in response to the growing rate of antibiotic resistant bacteria such as Staphylococcus aureus, Acinetobacter baumannii, and E. coli. As a tetracycline derivative antibiotic, its structural modifications has expanded its therapeutic activity to include Gram-positive and Gram-negative organisms, including those of multi-drug resistance.

Vancomycin-resistant Enterococcus, or vancomycin-resistant enterococci (VRE), are bacterial strains of the genus Enterococcus that are resistant to the antibiotic vancomycin.

Antibiotic sensitivity testing or antibiotic susceptibility testing is the measurement of the susceptibility of bacteria to antibiotics. It is used because bacteria may have resistance to some antibiotics. Sensitivity testing results can allow a clinician to change the choice of antibiotics from empiric therapy, which is when an antibiotic is selected based on clinical suspicion about the site of an infection and common causative bacteria, to directed therapy, in which the choice of antibiotic is based on knowledge of the organism and its sensitivities.
A drug of last resort (DoLR), also known as a heroic dose, is a pharmaceutical drug which is tried after all other drug options have failed to produce an adequate response in the patient. Drug resistance, such as antimicrobial resistance or antineoplastic resistance, may make the first-line drug ineffective, especially in case of multidrug-resistant pathogens and tumors. Such an alternative may be outside of extant regulatory requirements or medical best practices, in which case it may be viewed as salvage therapy.
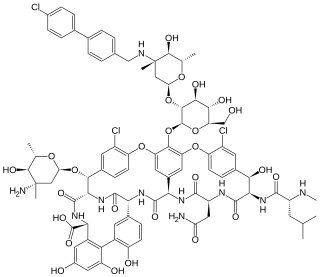
Oritavancin, sold under the brand name Orbactiv among others, is a semisynthetic glycopeptide antibiotic medication for the treatment of serious Gram-positive bacterial infections. Its chemical structure as a lipoglycopeptide is similar to vancomycin.

Cefoxitin is a second-generation cephamycin antibiotic developed by Merck & Co., Inc. from Cephamycin C in the year following its discovery, 1972. It was synthesized in order to create an antibiotic with a broader spectrum. It is often grouped with the second-generation cephalosporins. Cefoxitin requires a prescription and as of 2010 is sold under the brand name Mefoxin by Bioniche Pharma, LLC. The generic version of cefoxitin is known as cefoxitin sodium.
Streptogramins are a class of antibiotics.

Ceftobiprole, sold under the brand name Zevtera among others, is a fifth-generation cephalosporin antibacterial used for the treatment of hospital-acquired pneumonia and community-acquired pneumonia. It is marketed by Basilea Pharmaceutica under the brand names Zevtera and Mabelio. Like other cephalosporins, ceftobiprole exerts its antibacterial activity by binding to important penicillin-binding proteins and inhibiting their transpeptidase activity which is essential for the synthesis of bacterial cell walls. Ceftobiprole has high affinity for penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus strains and retains its activity against strains that express divergent mecA gene homologues. Ceftobiprole also binds to penicillin-binding protein 2b in Streptococcus pneumoniae (penicillin-intermediate), to penicillin-binding protein 2x in Streptococcus pneumoniae (penicillin-resistant), and to penicillin-binding protein 5 in Enterococcus faecalis.

Dalbavancin, sold under the brand names Dalvance in the US and Xydalba in the EU among others, is a second-generation lipoglycopeptide antibiotic medication. It belongs to the same class as vancomycin, the most widely used and one of the treatments available to people infected with methicillin-resistant Staphylococcus aureus (MRSA).
mecA is a gene found in bacterial cells which allows them to be resistant to antibiotics such as methicillin, penicillin and other penicillin-like antibiotics.
Staphylococcus pseudintermedius is a gram-positive spherically shaped bacterium of the genus Staphylococcus found worldwide. It is primarily a pathogen for domestic animals, but has been known to affect humans as well. S. pseudintermedius is an opportunistic pathogen that secretes immune-modulating virulence factors, has many adhesion factors, and the potential to create biofilms, all of which help to determine the pathogenicity of the bacterium. Diagnoses of S. pseudintermedius have traditionally been made using cytology, plating, and biochemical tests. More recently, molecular technologies like MALDI-TOF, DNA hybridization and PCR have become preferred over biochemical tests for their more rapid and accurate identifications. This includes the identification and diagnosis of antibiotic resistant strains.
ESKAPE is an acronym comprising the scientific names of six highly virulent and antibiotic resistant bacterial pathogens including: Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp. The acronym is sometimes extended to ESKAPEE to include Escherichia coli. This group of Gram-positive and Gram-negative bacteria can evade or 'escape' commonly used antibiotics due to their increasing multi-drug resistance (MDR). As a result, throughout the world, they are the major cause of life-threatening nosocomial or hospital-acquired infections in immunocompromised and critically ill patients who are most at risk. P. aeruginosa and S. aureus are some of the most ubiquitous pathogens in biofilms found in healthcare. P. aeruginosa is a Gram-negative, rod-shaped bacterium, commonly found in the gut flora, soil, and water that can be spread directly or indirectly to patients in healthcare settings. The pathogen can also be spread in other locations through contamination, including surfaces, equipment, and hands. The opportunistic pathogen can cause hospitalized patients to have infections in the lungs, blood, urinary tract, and in other body regions after surgery. S. aureus is a Gram-positive, cocci-shaped bacterium, residing in the environment and on the skin and nose of many healthy individuals. The bacterium can cause skin and bone infections, pneumonia, and other types of potentially serious infections if it enters the body. S. aureus has also gained resistance to many antibiotic treatments, making healing difficult. Because of natural and unnatural selective pressures and factors, antibiotic resistance in bacteria usually emerges through genetic mutation or acquires antibiotic-resistant genes (ARGs) through horizontal gene transfer - a genetic exchange process by which antibiotic resistance can spread.
Kerry L. LaPlante is an American pharmacist, academic and researcher. She is the Dean at the University of Rhode Island College of Pharmacy. She is a Professor of Pharmacy and former department Chair of the Department of Pharmacy Practice at the University of Rhode Island, an adjunct professor of medicine at Brown University, an Infectious Diseases Pharmacotherapy Specialist, and the Director of the Rhode Island Infectious Diseases Fellowship and Research Programs at the Veterans Affairs Medical Center in Providence, Rhode Island.















