
Staphylococcus aureus is a Gram-positive spherically shaped bacterium, a member of the Bacillota, and is a usual member of the microbiota of the body, frequently found in the upper respiratory tract and on the skin. It is often positive for catalase and nitrate reduction and is a facultative anaerobe that can grow without the need for oxygen. Although S. aureus usually acts as a commensal of the human microbiota, it can also become an opportunistic pathogen, being a common cause of skin infections including abscesses, respiratory infections such as sinusitis, and food poisoning. Pathogenic strains often promote infections by producing virulence factors such as potent protein toxins, and the expression of a cell-surface protein that binds and inactivates antibodies. S. aureus is one of the leading pathogens for deaths associated with antimicrobial resistance and the emergence of antibiotic-resistant strains, such as methicillin-resistant S. aureus (MRSA), is a worldwide problem in clinical medicine. Despite much research and development, no vaccine for S. aureus has been approved.

Vancomycin-resistant Staphylococcus aureus (VRSA) are strains of Staphylococcus aureus that have acquired resistance to the glycopeptide antibiotic vancomycin. Bacteria can acquire resistant genes either by random mutation or through the transfer of DNA from one bacterium to another. Resistance genes interfere with the normal antibiotic function and allow a bacteria to grow in the presence of the antibiotic. Resistance in VRSA is conferred by the plasmid-mediated vanA gene and operon. Although VRSA infections are uncommon, VRSA is often resistant to other types of antibiotics and a potential threat to public health because treatment options are limited. VRSA is resistant to many of the standard drugs used to treat S. aureus infections. Furthermore, resistance can be transferred from one bacterium to another.

Staphylococcus haemolyticus is a member of the coagulase-negative staphylococci (CoNS). It is part of the skin flora of humans, and its largest populations are usually found at the axillae, perineum, and inguinal areas. S. haemolyticus also colonizes primates and domestic animals. It is a well-known opportunistic pathogen, and is the second-most frequently isolated CoNS. Infections can be localized or systemic, and are often associated with the insertion of medical devices. The highly antibiotic-resistant phenotype and ability to form biofilms make S. haemolyticus a difficult pathogen to treat. Its most closely related species is Staphylococcus borealis.

Staphylococcus epidermidis is a Gram-positive bacterium, and one of over 40 species belonging to the genus Staphylococcus. It is part of the normal human microbiota, typically the skin microbiota, and less commonly the mucosal microbiota and also found in marine sponges. It is a facultative anaerobic bacteria. Although S. epidermidis is not usually pathogenic, patients with compromised immune systems are at risk of developing infection. These infections are generally hospital-acquired. S. epidermidis is a particular concern for people with catheters or other surgical implants because it is known to form biofilms that grow on these devices. Being part of the normal skin microbiota, S. epidermidis is a frequent contaminant of specimens sent to the diagnostic laboratory.

Protein A is a 42 kDa surface protein originally found in the cell wall of the bacteria Staphylococcus aureus. It is encoded by the spa gene and its regulation is controlled by DNA topology, cellular osmolarity, and a two-component system called ArlS-ArlR. It has found use in biochemical research because of its ability to bind immunoglobulins. It is composed of five homologous Ig-binding domains that fold into a three-helix bundle. Each domain is able to bind proteins from many mammalian species, most notably IgGs. It binds the heavy chain within the Fc region of most immunoglobulins and also within the Fab region in the case of the human VH3 family. Through these interactions in serum, where IgG molecules are bound in the wrong orientation, the bacteria disrupts opsonization and phagocytosis.
mecA is a gene found in bacterial cells which allows them to be resistant to antibiotics such as methicillin, penicillin and other penicillin-like antibiotics.

Staphylococcus capitis is a coagulase-negative species (CoNS) of Staphylococcus. It is part of the normal flora of the skin of the human scalp, face, neck, scrotum, and ears and has been associated with prosthetic valve endocarditis, but is rarely associated with native valve infection.
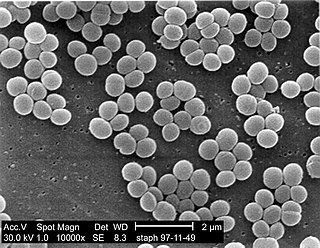
Staphylococcus is a genus of Gram-positive bacteria in the family Staphylococcaceae from the order Bacillales. Under the microscope, they appear spherical (cocci), and form in grape-like clusters. Staphylococcus species are facultative anaerobic organisms.

Alkalihalobacillus clausii or its old scientific name Bacillus clausii is a rod-shaped, motile, and spore-forming bacterium that lives in the soil but is also a natural microflora of the mammalian gastrointestinal tract. It is classified as probiotic microorganism that maintains a symbiotic relationship with the host organism. It is currently being studied in relation to respiratory infections and some gastrointestinal disorders. Bacillus clausii has been found to produce antimicrobial substances that are active against gram-positive bacteria including Staphylococcus aureus, Enterococcus faecium and Clostridium difficile. It is sold as an anti-diarrhoea and under the brand name Erceflora by Sanofi.
Bacterial small RNAs (bsRNA) are small RNAs produced by bacteria; they are 50- to 500-nucleotide non-coding RNA molecules, highly structured and containing several stem-loops. Numerous sRNAs have been identified using both computational analysis and laboratory-based techniques such as Northern blotting, microarrays and RNA-Seq in a number of bacterial species including Escherichia coli, the model pathogen Salmonella, the nitrogen-fixing alphaproteobacterium Sinorhizobium meliloti, marine cyanobacteria, Francisella tularensis, Streptococcus pyogenes, the pathogen Staphylococcus aureus, and the plant pathogen Xanthomonas oryzae pathovar oryzae. Bacterial sRNAs affect how genes are expressed within bacterial cells via interaction with mRNA or protein, and thus can affect a variety of bacterial functions like metabolism, virulence, environmental stress response, and structure.
Rsa RNAs are non-coding RNAs found in the bacterium Staphylococcus aureus. The shared name comes from their discovery, and does not imply homology. Bioinformatics scans identified the 16 Rsa RNA families named RsaA-K and RsaOA-OG. Others, RsaOH-OX, were found thanks to an RNomic approach. Although the RNAs showed varying expression patterns, many of the newly discovered RNAs were shown to be Hfq-independent and most carried a C-rich motif (UCCC).
Staphylococcus cohnii is a Gram-positive, coagulase-negative member of the bacterial genus Staphylococcus consisting of clustered cocci. The species commonly lives on human skin; clinical isolates have shown high levels of antibiotic resistance. A strain of S. cohnii was found to contain a mobile genetic element very similar to the staphylococcal cassette chromosome encoding methicillin resistance element seen in Staphylococcus aureus.
Staphylococcus rostri is a Gram-positive, coagulase-negative member of the bacterial genus Staphylococcus consisting of clustered cocci. This species was originally isolated from the noses of healthy pigs; the name is derived from the Latin rostrum or "the snout of a swine".
Staphylococcus delphini is a Gram-positive, coagulase-positive member of the bacterial genus Staphylococcus consisting of single, paired, and clustered cocci. Strains of this species were originally isolated from aquarium-raised dolphins suffering from skin lesions.
Staphylococcus equorum is a gram-positive, coagulase-negative member of the bacterial genus Staphylococcus consisting of clustered cocci. Originally isolated from the skin of healthy horses, this species contains a cell wall similar to that of Staphylococcus xylosus.

Staphylococcus hyicus is a Gram-positive, facultatively anaerobic bacterium in the genus Staphylococcus. It consists of clustered cocci and forms white circular colonies when grown on blood agar. S. hyicus is a known animal pathogen. It causes disease in poultry, cattle, horses, and pigs. Most notably, it is the agent that causes porcine exudative epidermitis, also known as greasy pig disease, in piglets. S. hyicus is generally considered to not be zoonotic, however it has been shown to be able to cause bacteremia and sepsis in humans.
Staphylococcus intermedius is a Gram-positive, catalase positive member of the bacterial genus Staphylococcus consisting of clustered cocci. Strains of this species were originally isolated from the anterior nares of pigeons, dogs, cats, mink, and horses. Many of the isolated strains show coagulase activity. Clinical tests for detection of methicillin-resistant S. aureus may produce false positives by detecting S. intermedius, as this species shares some phenotypic traits with methicillin-resistant S. aureus strains. It has been theorized that S. intermedius has previously been misidentified as S. aureus in human dog bite wound infections, which is why molecular technologies such as MALDI-TOF and PCR are preferred in modern veterinary clinical microbiology laboratories for their more accurate identifications over biochemical tests. S. intermedius is largely phenotypically indiscriminate from Staphylococcus pseudintermedius and Staphylococcus delphini, and therefore the three organisms are considered to be included in the more general 'Staphylococcus intermedius group'.
Staphylococcus schleiferi is a Gram-positive, cocci-shaped bacterium of the family Staphylococcaceae. It is facultatively anaerobic, coagulase-variable, and can be readily cultured on blood agar where the bacterium tends to form opaque, non-pigmented colonies and beta (β) hemolysis. There exists two subspecies under the species S. schleiferi: Staphylococcus schleiferi subsp. schleiferi and Staphylococcus schleiferi subsp. coagulans.
Staphylococcus pseudintermedius is a gram positive coccus bacteria of the genus Staphylococcus found worldwide. It is primarily a pathogen for domestic animals, but has been known to affect humans as well. S. pseudintermedius is an opportunistic pathogen that secretes immune modulating virulence factors, has many adhesion factors, and the potential to create biofilms, all of which help to determine the pathogenicity of the bacterium. Diagnoses of Staphylococcus pseudintermedius have traditionally been made using cytology, plating, and biochemical tests. More recently, molecular technologies like MALDI-TOF, DNA hybridization and PCR have become preferred over biochemical tests for their more rapid and accurate identifications. This includes the identification and diagnosis of antibiotic resistant strains.
Aeromonas simiae is a Gram-negative, oxidase- and catalase-positive motile bacterium of the genus Aeromonas, with a polar flagellum, isolated from the faeces of a healthy monkey.








